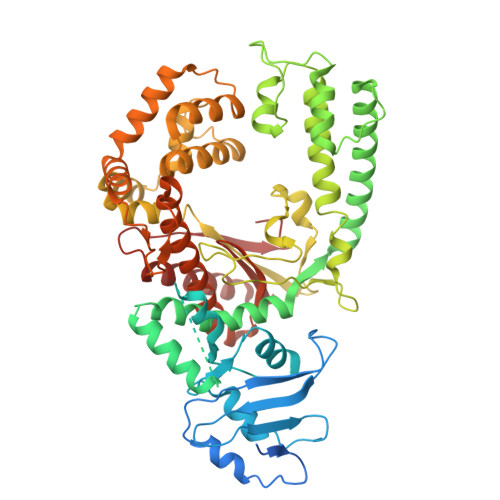
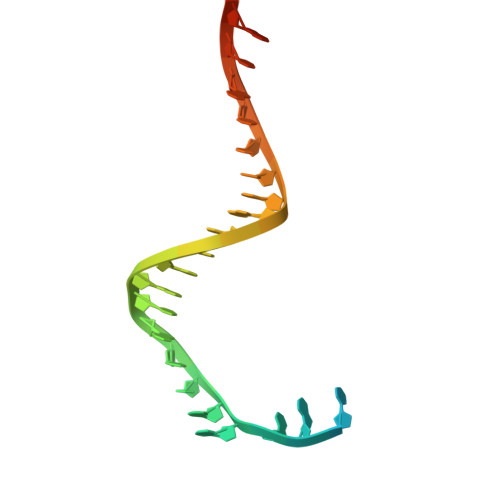
Cryo-EM Structures of the Plasmodium falciparum Apicoplast DNA Polymerase.
Lo, C.Y., Ung, A.R., Koley, T., Nelson, S.W., Gao, Y.(2024) J Mol Biology 436: 168842-168842
- PubMed: 39490679
- DOI: https://doi.org/10.1016/j.jmb.2024.168842
- Primary Citation of Related Structures:
9CAY, 9DG1 - PubMed Abstract:
The apicoplast DNA polymerase (apPol) from Plasmodium falciparum is essential for the parasite's survival, making it a prime target for antimalarial therapies. Here, we present cryo-electron microscopy structures of the apPol in complex with DNA and incoming nucleotide, offering insights into its molecular mechanisms. Our structural analysis reveals that apPol contains critical residues for high-fidelity DNA synthesis, but lacks certain structural elements to confer processive DNA synthesis during replication, suggesting the presence of additional accessory factors. The enzyme exhibits large-scale conformational changes transitioning upon DNA and nucleotide binding, particularly within the fingers and thumb subdomains. These movements reveal potential allosteric sites that could serve as targets for drug design. Our findings provide a foundation for advancing the understanding of apPol's unique functional mechanisms and potentially offering new avenues for the development of novel inhibitors and therapeutic interventions against malaria.
- Department of BioSciences, Rice University, Houston, TX, 77005, USA.
Organizational Affiliation: