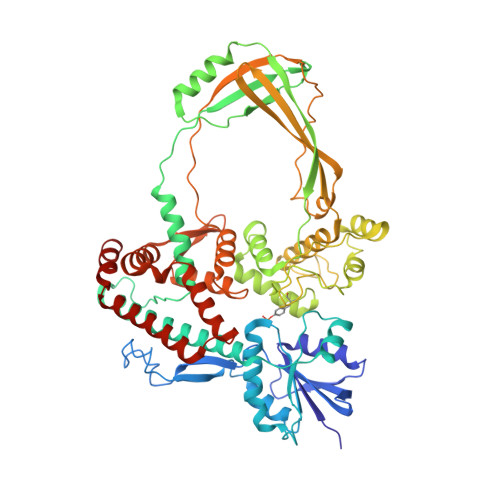
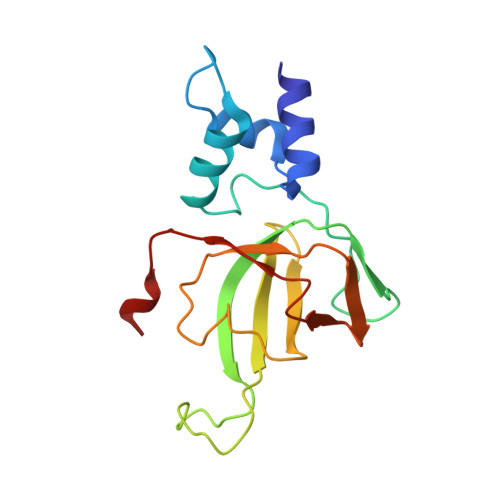
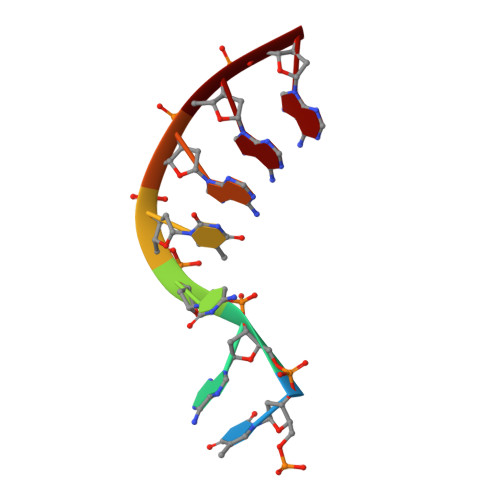
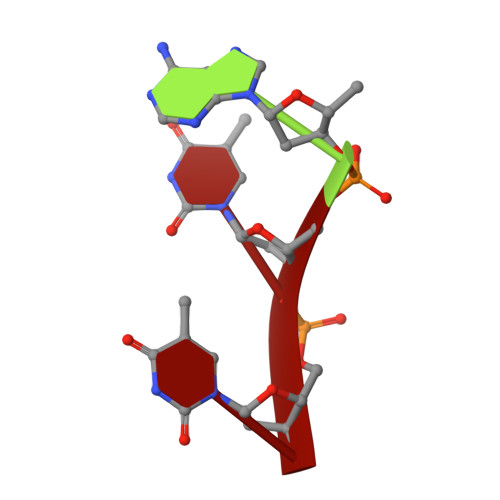
Structural insights into human topoisomerase 3 beta DNA and RNA catalysis and nucleic acid gate dynamics.
Yang, X., Chen, X., Yang, W., Pommier, Y.(2025) Nat Commun 16: 834-834
- PubMed: 39828754
- DOI: https://doi.org/10.1038/s41467-025-55959-y
- Primary Citation of Related Structures:
9C9W, 9C9Y, 9CA0, 9CA1, 9CA4, 9CAG, 9CAH, 9CAJ, 9CAK, 9CAL - PubMed Abstract:
Type IA topoisomerases (TopoIAs) are present in all living organisms. They resolve DNA/RNA catenanes, knots and supercoils by breaking and rejoining single-stranded DNA/RNA segments and allowing the passage of another nucleic acid segment through the break. Topoisomerase III-β (TOP3B), the only RNA topoisomerase in metazoans, promotes R-loop disassembly and translation of mRNAs. Defects in TOP3B lead to severe neurological diseases. We present a series of cryo-EM structures of human TOP3B with its cofactor TDRD3 during cleavage and rejoining of DNA or RNA, thus elucidating the roles of divalent metal ions and key enzyme residues in each step of the catalytic cycle. We also obtained the structure of an open-gate configuration that addresses the long-standing question of the strand-passage mechanism. Our studies reveal how TOP3B catalyzes both DNA and RNA relaxation, while TOP3A acts only on DNA.
- Developmental Therapeutics Branch & Laboratory of Molecular Pharmacology, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD, USA.
Organizational Affiliation: