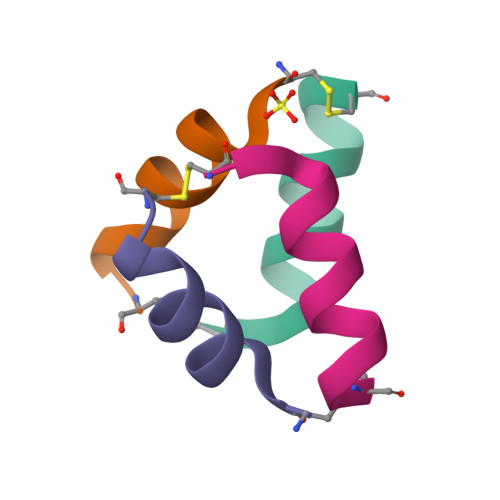
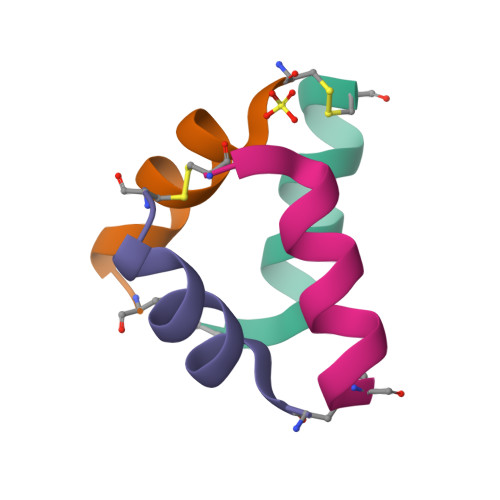
Structural Characterization of Disulfide-Linked p53-Derived Peptide Dimers.
DiGiorno, M.C., Vithanage, N., Victorio, C.G., Kreitler, D.F., Outlaw, V.K., Sawyer, N.(2024) Res Sq
- PubMed: 39070635
- DOI: https://doi.org/10.21203/rs.3.rs-4644285/v1
- Primary Citation of Related Structures:
9C5S - PubMed Abstract:
Disulfide bonds provide a convenient method for chemoselective alteration of peptide and protein structure and function. We previously reported that mild oxidation of a p53-derived bisthiol peptide (CTFANLWRLLAQNC) under dilute non-denaturing conditions led to unexpected disulfide-linked dimers as the exclusive product. The dimers were antiparallel, significantly α-helical, resistant to protease degradation, and easily reduced back to the original bisthiol peptide. Here we examine the intrinsic factors influencing peptide dimerization using a combination of amino acid substitution, circular dichroism (CD) spectroscopy, and X-ray crystallography. CD analysis of peptide variants suggests critical roles for Leu6 and Leu10 in the formation of stable disulfide-linked dimers. The 1.0 Å resolution crystal structure of the peptide dimer supports these data, revealing a leucine-rich LxxLL dimer interface with canonical knobs-into-holes packing. Two levels of higher-order oligomerization are also observed in the crystal: an antiparallel "dimer of dimers" mediated by Phe3 and Trp7 residues in the asymmetric unit and a tetramer of dimers mediated by Trp7 and Leu10. In CD spectra of Trp-containing peptide variants, minima at 227 nm provide evidence for the dimer of dimers in dilute aqueous solution. Importantly, and in contrast to the original dimer model, the canonical leucine-rich core and robust dimerization of most peptide variants suggests a tunable molecular architecture to target various proteins and evaluate how folding and oligomerization impact various properties, such as cell permeability.