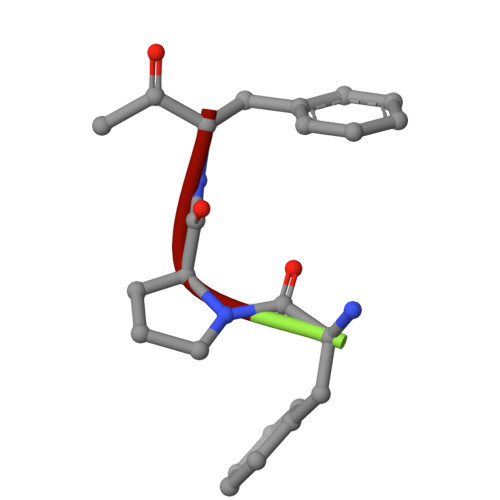
Replacement of a single residue changes the primary specificity of thrombin.
Dei Rossi, A., Deavila, S., Mohammed, B.M., Korolev, S., Di Cera, E.(2025) J Thromb Haemost 23: 1241-1246
- PubMed: 39756655
- DOI: https://doi.org/10.1016/j.jtha.2024.12.024
- Primary Citation of Related Structures:
9C50 - PubMed Abstract:
Thrombin prefers substrates carrying Arg at the site of cleavage (P1) because of the presence of D189 in the primary specificity (S1) pocket but can also cleave substrates carrying Phe at P1. The structural basis of this property is unknown. Solve the X-ray structure of thrombin bound to a ligand carrying Phe at P1 and investigate the effects of replacing D189. X-ray crystallography is used to solve the structure of thrombin bound to the irreversible inhibitor H-D-Phe-Pro-Phe-CH 2 Cl (PPPCK). Residue D189 is mutated to Ala, Lys, Phe and Ser. The X-ray structure of the thrombin-PPPCK complex is solved at 2.5 Å resolution and compared to the structure of thrombin bound to H-D-Phe-Pro-Arg-CH 2 Cl (PPACK). PPPCK binds to thrombin in a conformation similar to that of PPACK, but Phe at P1 makes no contacts with D189. Replacement of D189 with Ala, Lys, Phe or Ser reverses both substrate preference and stability enhancement from Arg to Phe. D189 in the S1 pocket confers thrombin "trypsin-like" specificity for Arg at P1. However, the S1 pocket is wide enough to also enable "chymotrypsin-like" specificity for Phe at P1. Consistent with these structural features, a single amino acid replacement (D189A) switches thrombin specificity from trypsin-like to chymotrypsin-like, converting the substrate preference from H-D-Phe-Pro-Arg-p-nitroanilide to H-D-Phe-Pro-Phe-p-nitroanilide and preferential stability enhancement from PPACK to PPPCK. The observation that thrombin specificity is controlled mainly by a single residue establishes a new paradigm in the field of trypsin-like proteases.
- Edward A. Doisy Department of Biochemistry and Molecular Biology, Saint Louis University School of Medicine, St. Louis, MO 63104 USA.
Organizational Affiliation: