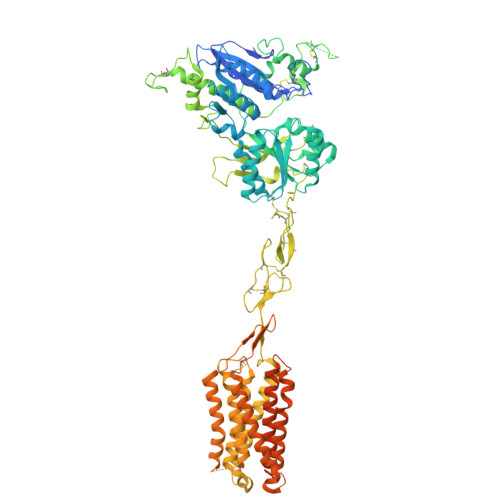
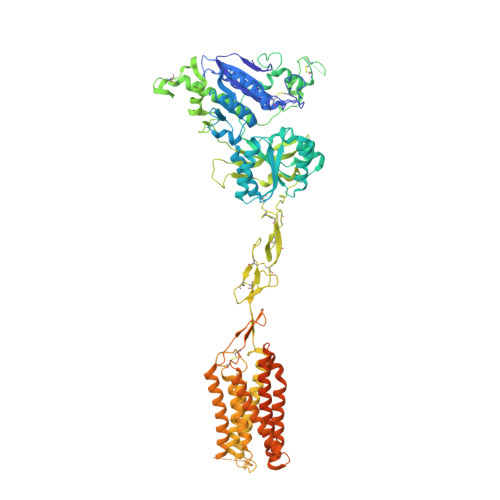
Large library docking identifies positive allosteric modulators of the calcium-sensing receptor.
Liu, F., Wu, C.G., Tu, C.L., Glenn, I., Meyerowitz, J., Kaplan, A.L., Lyu, J., Cheng, Z., Tarkhanova, O.O., Moroz, Y.S., Irwin, J.J., Chang, W., Shoichet, B.K., Skiniotis, G.(2024) Science 385: eado1868-eado1868
- PubMed: 39298584
- DOI: https://doi.org/10.1126/science.ado1868
- Primary Citation of Related Structures:
9C1P, 9C2F - PubMed Abstract:
Positive allosteric modulator (PAM) drugs enhance the activation of the calcium-sensing receptor (CaSR) and suppress parathyroid hormone (PTH) secretion. Unfortunately, these hyperparathyroidism-treating drugs can induce hypocalcemia and arrhythmias. Seeking improved modulators, we docked libraries of 2.7 million and 1.2 billion molecules against the CaSR structure. The billion-molecule docking found PAMs with a 2.7-fold higher hit rate than the million-molecule library, with hits up to 37-fold more potent. Structure-based optimization led to nanomolar leads. In ex vivo organ assays, one of these PAMs was 100-fold more potent than the standard of care, cinacalcet, and reduced serum PTH levels in mice without the hypocalcemia typical of CaSR drugs. As determined from cryo-electron microscopy structures, the PAMs identified here promote CaSR conformations that more closely resemble the activated state than those induced by the established drugs.
- Department of Pharmaceutical Chemistry, University of California, San Francisco, San Francisco, CA 94143, USA.
Organizational Affiliation: