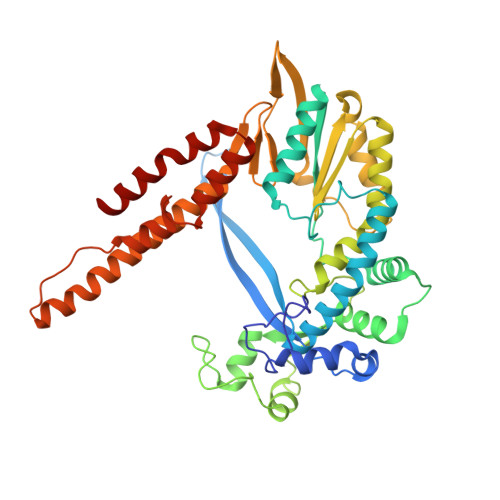
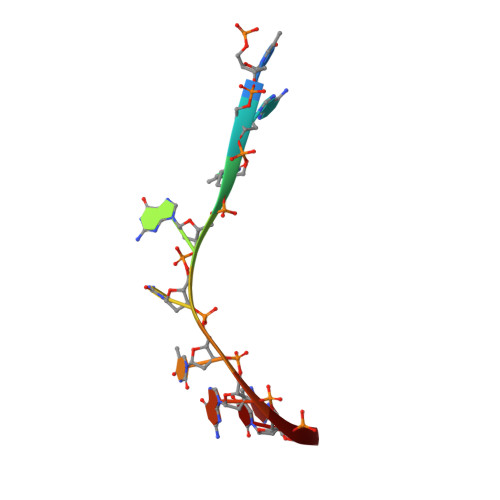
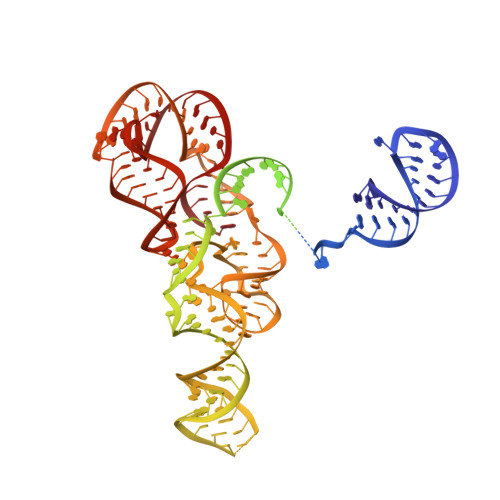
Phage-triggered reverse transcription assembles a toxic repetitive gene from a noncoding RNA.
Wilkinson, M.E., Li, D., Gao, A., Macrae, R.K., Zhang, F.(2024) Science 386: eadq3977-eadq3977
- PubMed: 39208082
- DOI: https://doi.org/10.1126/science.adq3977
- Primary Citation of Related Structures:
9C0I, 9C0J - PubMed Abstract:
Reverse transcription has frequently been co-opted for cellular functions and in prokaryotes is associated with protection against viral infection, but the underlying mechanisms of defense are generally unknown. Here, we show that in the DRT2 defense system the reverse transcriptase binds a neighboring pseudoknotted noncoding RNA. Upon bacteriophage infection, a template region of this RNA is reverse transcribed into an array of tandem repeats that reconstitute a promoter and open reading frame, allowing expression of a toxic repetitive protein and an abortive infection response. Biochemical reconstitution of this activity and cryogenic electron microscopy provide a molecular basis for repeat synthesis. Gene synthesis from a noncoding RNA is a new mode of genetic regulation in prokaryotes.
- Howard Hughes Medical Institute, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
Organizational Affiliation: