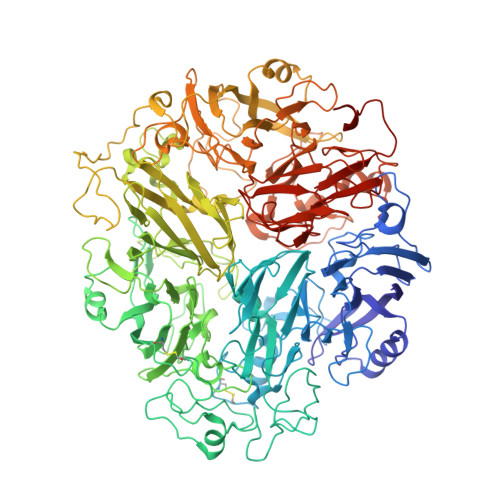
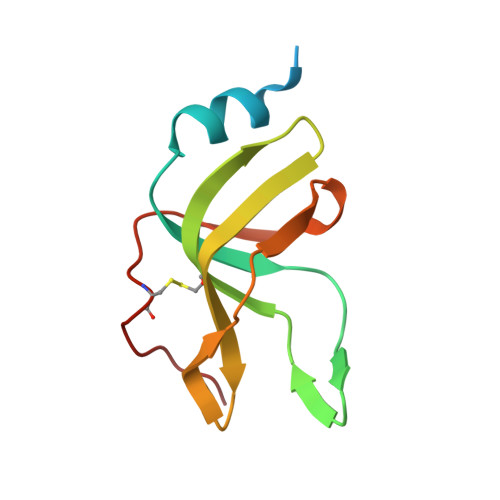
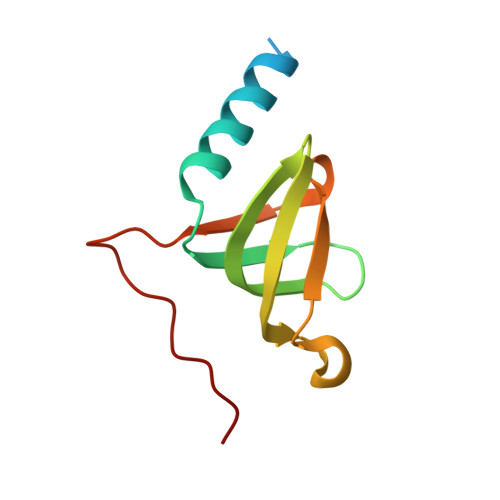
Cryo-EM Structure of the Mnx Protein Complex Reveals a Tunnel Framework for the Mechanism of Manganese Biomineralization.
Novikova, I.V., Soldatova, A.V., Moser, T.H., Thibert, S.M., Romano, C.A., Zhou, M., Tebo, B.M., Evans, J.E., Spiro, T.G.(2024) J Am Chem Soc 146: 22950-22958
- PubMed: 39056168
- DOI: https://doi.org/10.1021/jacs.3c06537
- Primary Citation of Related Structures:
9BXA - PubMed Abstract:
The global manganese cycle relies on microbes to oxidize soluble Mn(II) to insoluble Mn(IV) oxides. Some microbes require peroxide or superoxide as oxidants, but others can use O 2 directly, via multicopper oxidase (MCO) enzymes. One of these, MnxG from Bacillus sp. strain PL-12, was isolated in tight association with small accessory proteins, MnxE and MnxF. The protein complex, called Mnx, has eluded crystallization efforts, but we now report the 3D structure of a point mutant using cryo-EM single particle analysis, cross-linking mass spectrometry, and AlphaFold Multimer prediction. The β-sheet-rich complex features MnxG enzyme, capped by a heterohexameric ring of alternating MnxE and MnxF subunits, and a tunnel that runs through MnxG and its MnxE 3 F 3 cap. The tunnel dimensions and charges can accommodate the mechanistically inferred binuclear manganese intermediates. Comparison with the Fe(II)-oxidizing MCO, ceruloplasmin, identifies likely coordinating groups for the Mn(II) substrate, at the entrance to the tunnel. Thus, the 3D structure provides a rationale for the established manganese oxidase mechanism, and a platform for further experiments to elucidate mechanistic details of manganese biomineralization.
- Environmental Molecular Sciences Laboratory, Pacific Northwest National Laboratory, 3335 Innovation Blvd, Richland, Washington 99354, United States.
Organizational Affiliation: