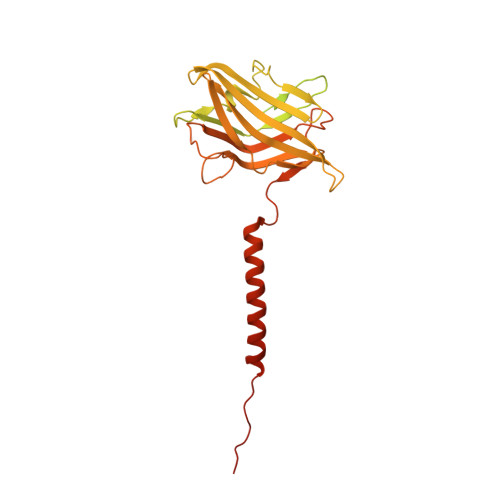
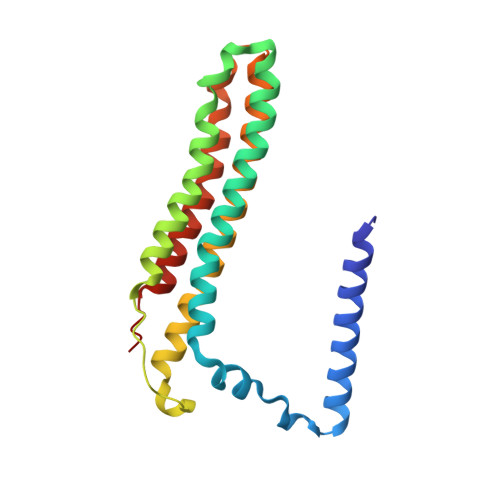
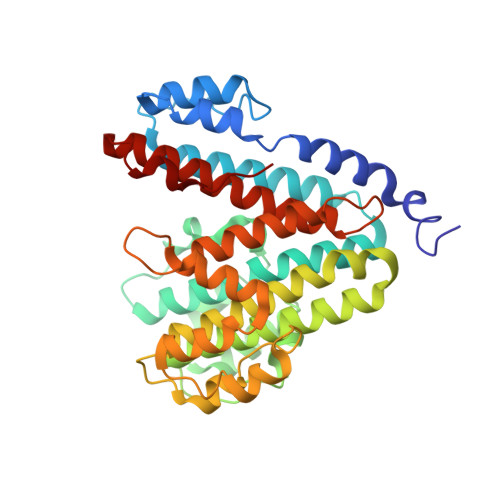
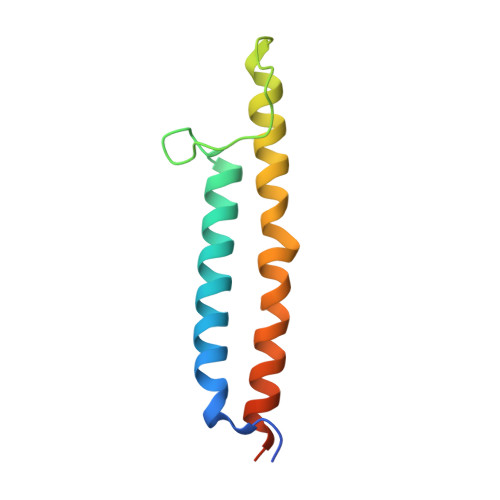
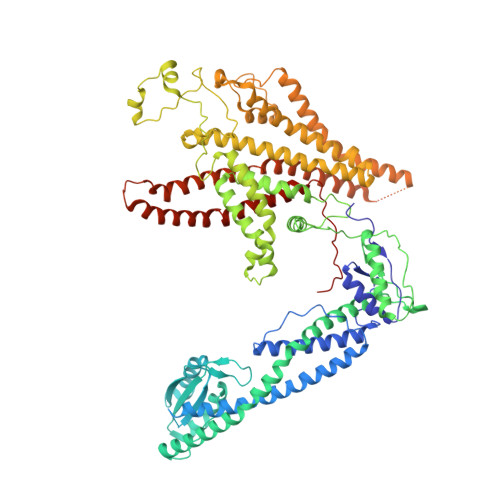
Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Wang, C., Jiang, W., Leitz, J., Yang, K., Esquivies, L., Wang, X., Shen, X., Held, R.G., Adams, D.J., Basta, T., Hampton, L., Jian, R., Jiang, L., Stowell, M.H.B., Baumeister, W., Guo, Q., Brunger, A.T.(2024) Nature 631: 899-904
- PubMed: 38838737
- DOI: https://doi.org/10.1038/s41586-024-07610-x
- Primary Citation of Related Structures:
9BRA, 9BRQ, 9BRR, 9BRS, 9BRT, 9BRU, 9BRY, 9BRZ - PubMed Abstract:
Synaptic vesicles are organelles with a precisely defined protein and lipid composition 1,2 , yet the molecular mechanisms for the biogenesis of synaptic vesicles are mainly unknown. Here, we discovered a well-defined interface between the synaptic vesicle V-ATPase and synaptophysin by in situ cryo-electron tomography and single particle cryo-electron microscopy of functional synaptic vesicles isolated from mouse brains 3 . The synaptic vesicle V-ATPase is an ATP-dependent proton pump that establishes the protein gradient across the synaptic vesicle, which in turn drives the uptake of neurotransmitters 4,5 . Synaptophysin 6 and its paralogs synaptoporin 7 and synaptogyrin 8 belong to a family of abundant synaptic vesicle proteins whose function is still unclear. We performed structural and functional studies of synaptophysin knockout mice, confirming the identity of synaptophysin as an interaction partner with the V-ATPase. Although there is little change in the conformation of the V-ATPase upon interaction with synaptophysin, the presence of synaptophysin in synaptic vesicles profoundly affects the copy number of V-ATPases. This effect on the topography of synaptic vesicles suggests that synaptophysin assists in their biogenesis. In support of this model, we observed that synaptophysin knockout mice exhibit severe seizure susceptibility, suggesting an imbalance of neurotransmitter release as a physiological consequence of the absence of synaptophysin.
- Department of Molecular and Cellular Physiology, Stanford University, Stanford, California, USA.
Organizational Affiliation: