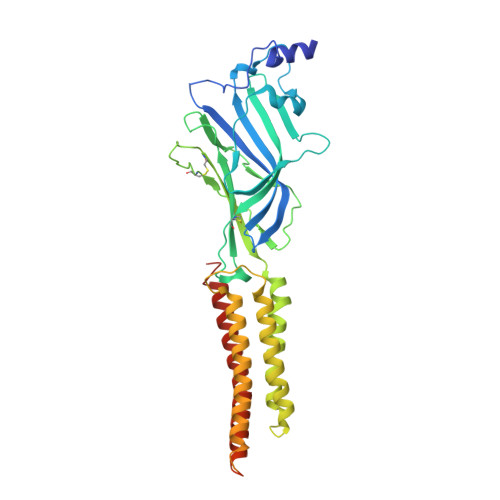
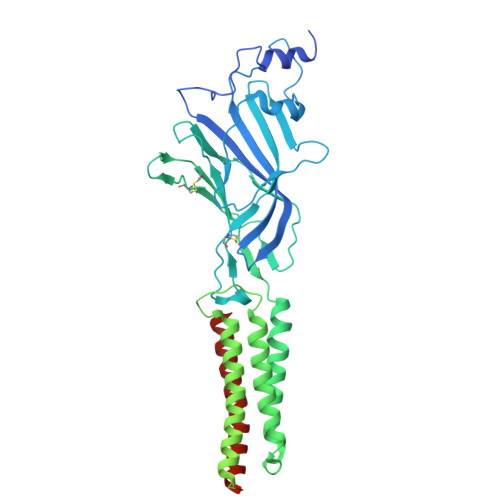
Mechanism of human alpha 3 beta GlyR regulation by intracellular M3/M4 loop phosphorylation and 2,6-di-tert-butylphenol interaction.
Liu, X., Krezel, M., Wang, W.(2025) Nat Commun 16: 5242-5242
- PubMed: 40473619
- DOI: https://doi.org/10.1038/s41467-025-60516-8
- Primary Citation of Related Structures:
9BOY, 9BOZ, 9BP7 - PubMed Abstract:
α3β glycine receptor (GlyR) is a subtype of GlyRs that belongs to the Cys-loop receptor superfamily. It is highly expressed in the spinal dorsal horn where sensory information is integrated. Under inflammatory conditions, the large unstructured intracellular M3/M4 loops of the α3 subunit are phosphorylated through the prostaglandin E2 (PGE 2 ) pathway, inhibiting ion conduction, and resulting in elevated pain sensation. A small molecule analgesic analog, 2,6-di-tert-butylphenol (2,6-DTBP) potentiates phosphorylated α3β GlyR through unclear mechanisms and relieves pain. Combining cryo-Electron Microscopy (cryo-EM) structures and single molecule Förster resonance energy transfer (smFRET) experiments, we show compaction of M3/M4 loop towards the ion conduction pore upon phosphorylation and further by 2,6-DTBP binding, which in turn modulates function through changing pore conformations and local electrostatics. We show that simultaneous interactions with the M3/M4 loop and the transmembrane domain (TM) is necessary for the potentiation of heteromeric α3β GlyR by 2,6-DTBP, while TM interaction alone is sufficient to potentiate homomeric α3 GlyR, explaining the mystery of why 2,6-DTBP potentiates only phosphorylated α3β GlyR. These findings show how post-translational modification of the unstructured intracellular M3/M4 loop may regulate Cys-loop receptor function, providing new perspectives in pain control and other pharmaceutical development targeting GlyRs and other Cys-loop receptors.
- Departments of Biophysics, University of Texas Southwestern Medical Center, Dallas, TX, USA.
Organizational Affiliation: