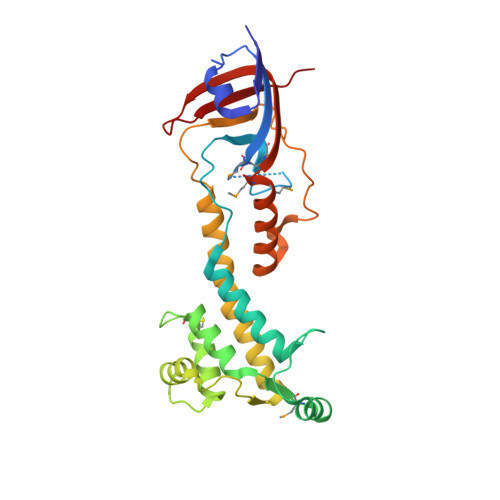
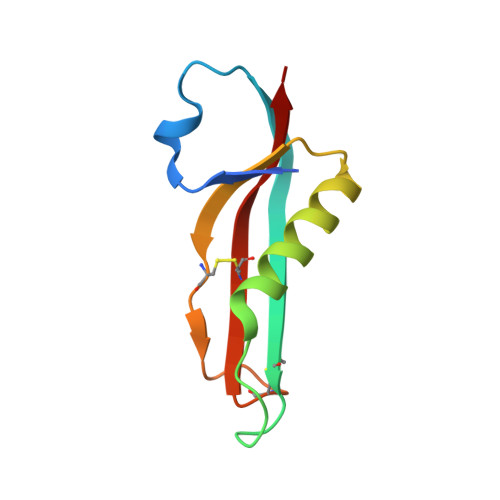
Molecular insights into the initiation step of the Rcs signaling pathway.
Watanabe, N., Savchenko, A.(2024) Structure 32: 1381
- PubMed: 38964336
- DOI: https://doi.org/10.1016/j.str.2024.06.003
- Primary Citation of Related Structures:
9BIY, 9BIZ, 9BJ0 - PubMed Abstract:
The Rcs pathway is repressed by the inner membrane protein IgaA under non-stressed conditions. This repression is hypothesized to be relieved by the binding of the outer membrane-anchored RcsF to IgaA. However, the precise mechanism by which RcsF binding triggers the signaling remains unclear. Here, we present the 1.8 Å resolution crystal structure capturing the interaction between IgaA and RcsF. Our comparative structural analysis, examining both the bound and unbound states of the periplasmic domain of IgaA (IgaAp), highlights rotational flexibility within IgaAp. Conversely, the conformation of RcsF remains unchanged upon binding. Our in vivo and in vitro studies do not support the model of a stable complex involving RcsF, IgaAp, and RcsDp. Instead, we demonstrate that the elements beyond IgaAp play a role in the interaction between IgaA and RcsD. These findings collectively allow us to propose a potential mechanism for the signaling across the inner membrane through IgaA.
- Department of Microbiology, Immunology and Infectious Diseases, University of Calgary, Calgary, AB, Canada; Center for Structural Biology for Infectious Diseases (CSBID) Chicago, IL, USA.
Organizational Affiliation: