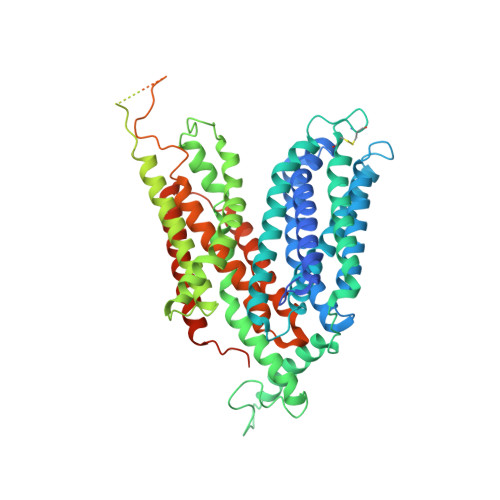
Structural basis for antibiotic transport and inhibition in PepT2.
Parker, J.L., Deme, J.C., Lichtinger, S.M., Kuteyi, G., Biggin, P.C., Lea, S.M., Newstead, S.(2024) Nat Commun 15: 8755-8755
- PubMed: 39384780
- DOI: https://doi.org/10.1038/s41467-024-53096-6
- Primary Citation of Related Structures:
9BIR, 9BIS, 9BIT, 9BIU - PubMed Abstract:
The uptake and elimination of beta-lactam antibiotics in the human body are facilitated by the proton-coupled peptide transporters PepT1 (SLC15A1) and PepT2 (SLC15A2). The mechanism by which SLC15 family transporters recognize and discriminate between different drug classes and dietary peptides remains unclear, hampering efforts to improve antibiotic pharmacokinetics through targeted drug design and delivery. Here, we present cryo-EM structures of the proton-coupled peptide transporter, PepT2 from Rattus norvegicus, in complex with the widely used beta-lactam antibiotics cefadroxil, amoxicillin and cloxacillin. Our structures, combined with pharmacophore mapping, molecular dynamics simulations and biochemical assays, establish the mechanism of beta-lactam antibiotic recognition and the important role of protonation in drug binding and transport.
- Department of Biochemistry, University of Oxford, Oxford, UK. joanne.parker@bioch.ox.ac.uk.
Organizational Affiliation: