Design of soluble HIV-1 envelope trimers free of covalent gp120-gp41 bonds with prevalent native-like conformation.
Zhang, P., Gorman, J., Tsybovsky, Y., Lu, M., Liu, Q., Gopan, V., Singh, M., Lin, Y., Miao, H., Seo, Y., Kwon, A., Olia, A.S., Chuang, G.Y., Geng, H., Lai, Y.T., Zhou, T., Mascola, J.R., Mothes, W., Kwong, P.D., Lusso, P.(2024) Cell Rep 43: 114518-114518
- PubMed: 39028623
- DOI: https://doi.org/10.1016/j.celrep.2024.114518
- Primary Citation of Related Structures:
9BER, 9BEW, 9BF6 - PubMed Abstract:
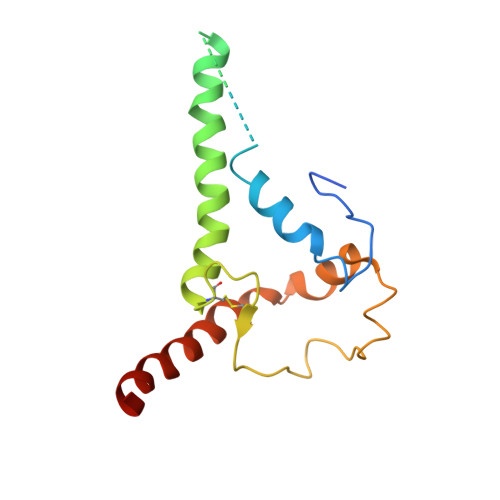
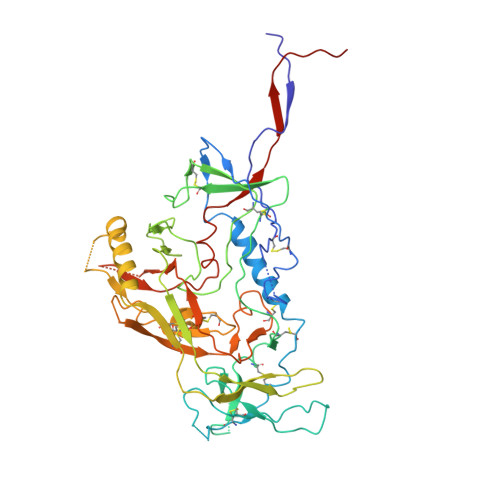
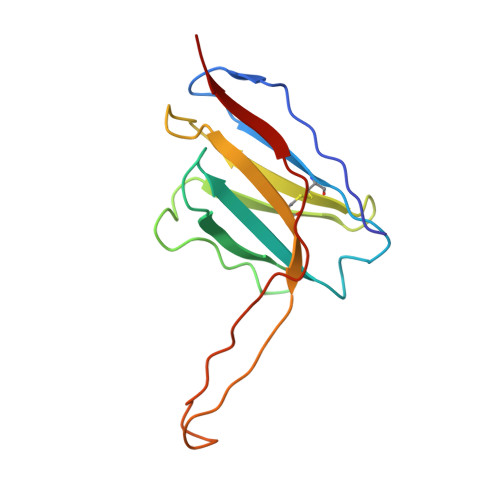
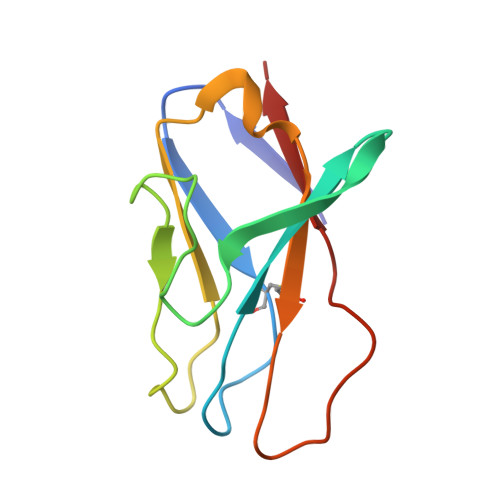
Soluble HIV-1 envelope (Env) trimers may serve as effective vaccine immunogens. The widely utilized SOSIP trimers have been paramount for structural studies, but the disulfide bond they feature between gp120 and gp41 constrains intersubunit mobility and may alter antigenicity. Here, we report an alternative strategy to generate stabilized soluble Env trimers free of covalent gp120-gp41 bonds. Stabilization was achieved by introducing an intrasubunit disulfide bond between the inner and outer domains of gp120, defined as interdomain lock (IDL). Correctly folded IDL trimers displaying a native-like antigenic profile were produced for HIV-1 Envs of different clades. Importantly, the IDL design abrogated CD4 binding while not affecting recognition by potent neutralizing antibodies to the CD4-binding site. By cryoelectron microscopy, IDL trimers were shown to adopt a closed prefusion configuration, while single-molecule fluorescence resonance energy transfer documented a high prevalence of native-like conformation. Thus, IDL trimers may be promising candidates as vaccine immunogens.
- Laboratory of Immunoregulation, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD 20892, USA. Electronic address: zhangp@ihm.ac.cn.
Organizational Affiliation: