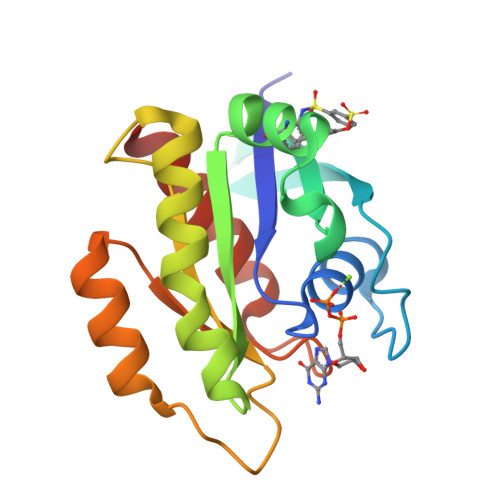
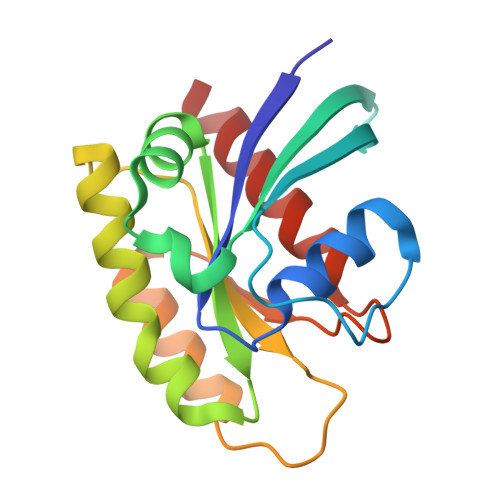
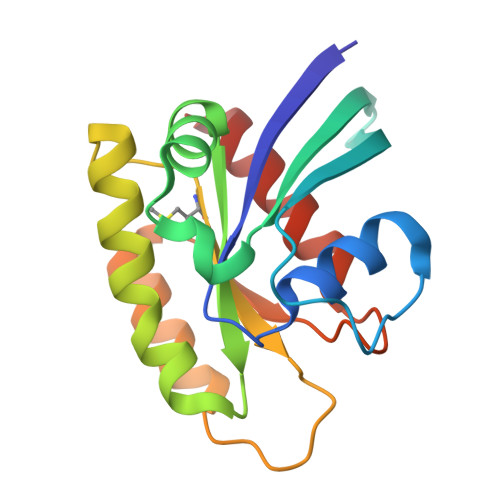
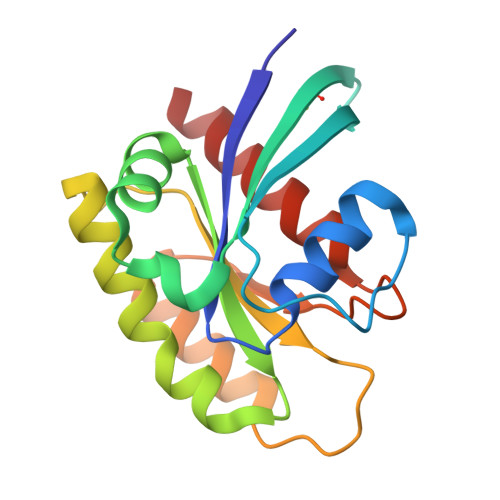
Small-Molecule KRAS Inhibitors by Tyrosine Covalent Bond Formation.
Landgraf, A., Brenner, R., Ghozayel, M., Bum-Erdene, K., Gonzalez-Gutierrez, G., Meroueh, S.(2025) ChemMedChem : e202400624-e202400624
- PubMed: 40099978
- DOI: https://doi.org/10.1002/cmdc.202400624
- Primary Citation of Related Structures:
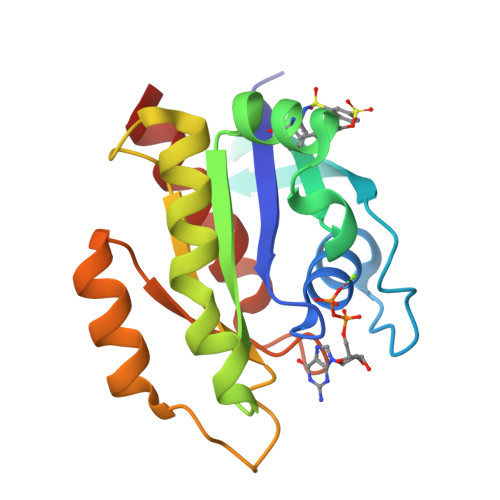
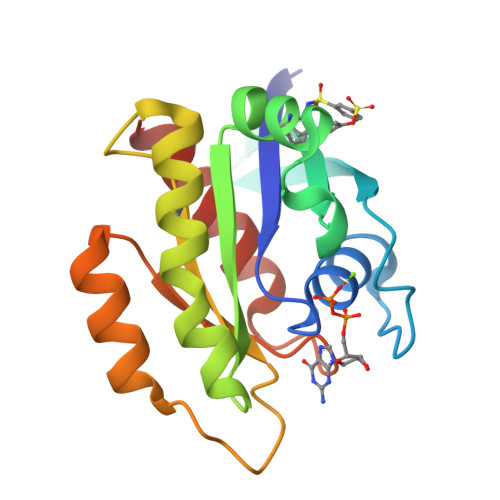
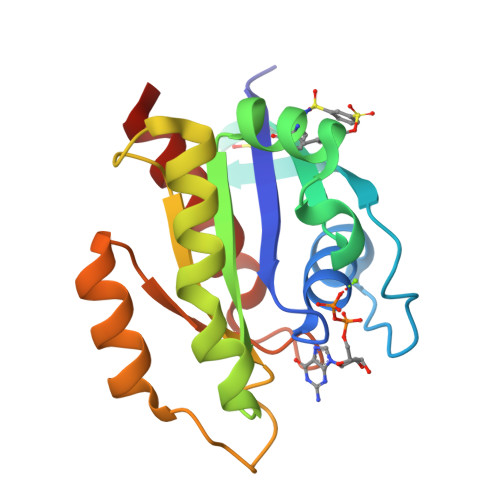
9BAI, 9BAJ, 9BAK - PubMed Abstract:
The development of the KRAS G12C inhibitor sotorasib was a major advance towards drugging KRAS. However, the G12C mutation is only found in about 10% of tumors with a KRAS mutation. KRAS tyrosine amino acids could provide alternative sites for covalent drug development. Here, we screen a library of aryl sulfonyl fluorides to explore whether tyrosines on KRAS are accessible for covalent bond formation. We identify compound 1 (SOF-436), which inhibits KRAS nucleotide exchange by guanine exchange factor SOS1 and the binding of KRAS to effector protein RAF. Tyr-64 was the major reaction site of 1 (SOF-436), although minor reaction at Tyr-71 was also observed. The fragment engages the Switch II pocket of KRAS based on mass spectrometry, nucleotide exchange, effector protein binding, nuclear magnetic resonance (NMR), and molecular dynamics simulations. Co-crystal structures of smaller fragments covalently bound to KRAS at Tyr-71 provide a strategy for the development of Switch I/II KRAS covalent inhibitors. A NanoBRET assay revealed that the compound and its analogs inhibit KRAS binding to RAF in mammalian cells. Although not yet suitable as chemical probes, these fragments provide starting points for small molecules to investigate tyrosine as a nucleophile for covalent inhibition of KRAS in tumors.
Organizational Affiliation:
Indiana University School of Medicine, Biochemistry, UNITED STATES OF AMERICA.