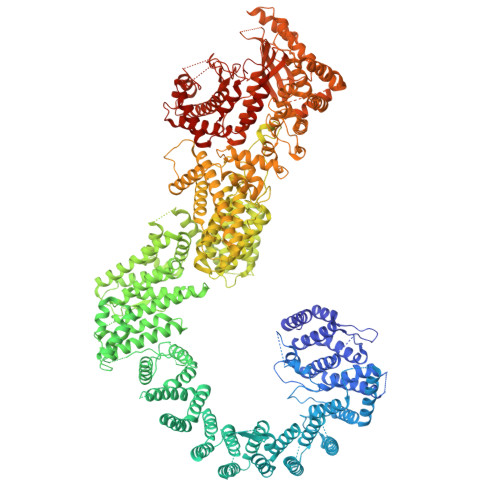
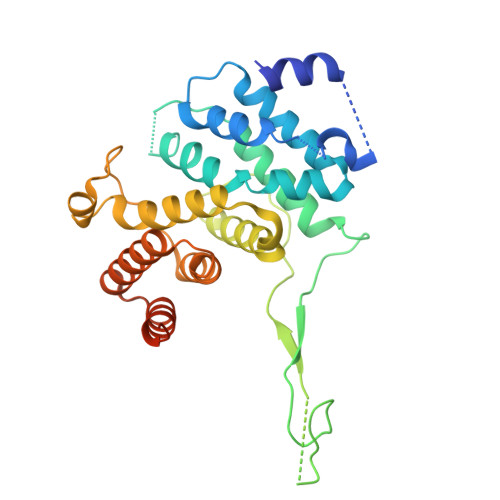
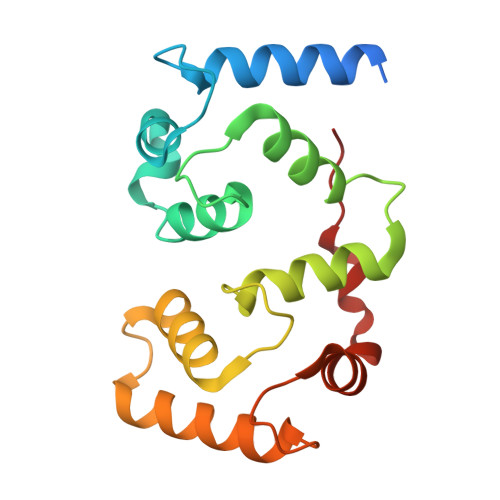
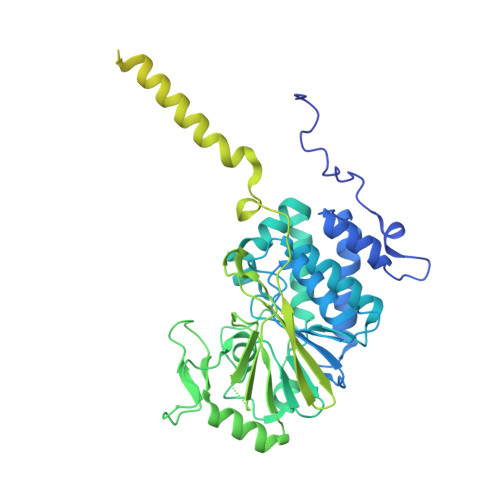
Structure of calcineurin bound to PI4KA reveals dual interface in both PI4KA and FAM126A.
Shaw, A.L., Suresh, S., Parson, M.A.H., Harris, N.J., Jenkins, M.L., Yip, C.K., Burke, J.E.(2024) Structure 32: 1973
- PubMed: 39216471
- DOI: https://doi.org/10.1016/j.str.2024.08.007
- Primary Citation of Related Structures:
9B9G - PubMed Abstract:
Phosphatidylinositol 4-kinase alpha (PI4KA) maintains the phosphatidylinositol 4-phosphate (PI4P) and phosphatidylserine pools of the plasma membrane. A key regulator of PI4KA is its association into a complex with TTC7 and FAM126 proteins. This complex can be regulated by the CNAβ1 isoform of the phosphatase calcineurin. We previously identified that CNAβ1 directly binds to FAM126A. Here, we report a cryoelectron microscopic (cryo-EM) structure of a truncated PI4KA complex bound to calcineurin, revealing a unique direct interaction between PI4KA and calcineurin. Hydrogen deuterium exchange mass spectrometry (HDX-MS) and computational analysis show that calcineurin forms a complex with an evolutionarily conserved IKISVT sequence in PI4KA's horn domain. We also characterized conserved LTLT and PSISIT calcineurin binding sequences in the C terminus of FAM126A. These dual sites in PI4KA and FAM126A are both in close proximity to phosphorylation sites in the PI4KA complex, suggesting key roles of calcineurin-regulated phosphosites in PI4KA regulation. This work reveals novel insight into how calcineurin can regulate PI4KA activity.
- Department of Biochemistry and Molecular Biology, The University of British Columbia, Vancouver, British Columbia V6T 1Z3, Canada; Department of Biochemistry and Microbiology, University of Victoria, Victoria, British Columbia V8W 2Y2, Canada.
Organizational Affiliation: