A nanobody-based therapeutic targeting Nipah virus limits viral escape.
Isaacs, A., Nieto, G.V., Zhang, X., Modhiran, N., Barr, J., Thakur, N., Low, Y.S., Parry, R.H., Barnes, J.B., Jara, R., Himelreichs, J., Yao, Y., Deride, C., Barthou-Gatica, B., Salinas-Rebolledo, C., Ehrenfeld, P., Hen, J.J., Hayes, N., Paramitha, D., Morgan, M.S., McMillan, C.L.D., Jones, M.L., Munro, T.P., Khromykh, A.A., Reading, P.C., Young, P.R., Chappell, K.J., Shi, Y., Bailey, D., Marsh, G.A., Chiu, S., Rojas-Fernandez, A., Watterson, D.(2025) Nat Struct Mol Biol 32: 1920-1931
- PubMed: 40629166
- DOI: https://doi.org/10.1038/s41594-025-01598-2
- Primary Citation of Related Structures:
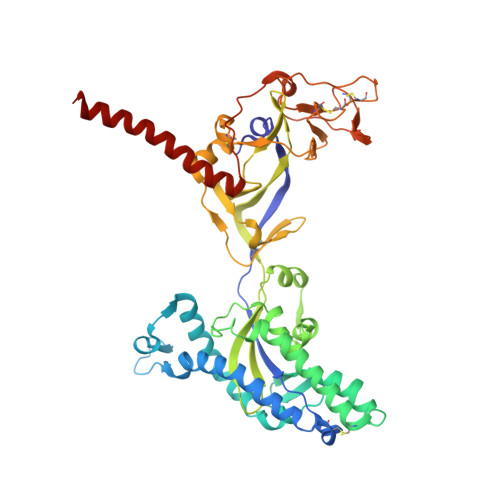
9B9E - PubMed Abstract:
Nipah virus (NiV) and Hendra virus (HeV) are highly pathogenic henipaviruses without approved human vaccines or therapies. Here, we report on a highly potent bispecific therapeutic that combines an anti-fusion glycoprotein nanobody with an anti-receptor-binding glycoprotein (RBP) antibody to deliver a dual-targeting biologic that is resistant to viral escape. We show that the nanobody, DS90, engages a unique, conserved site within the fusion glycoprotein of NiV and HeV and provides neutralization and complete protection from NiV disease. Bispecific engineering of DS90 with the anti-RBP monoclonal antibody m102.4 results in neutralization, elimination of viral escape and superior protection from NiV disease compared to leading monovalent approaches. These findings carry implications for the development of cross-neutralizing immunotherapies that limit the emergence of henipaviral escape mutants.
- School of Chemistry and Molecular Biosciences, The University of Queensland, Brisbane, Queensland, Australia.
Organizational Affiliation: