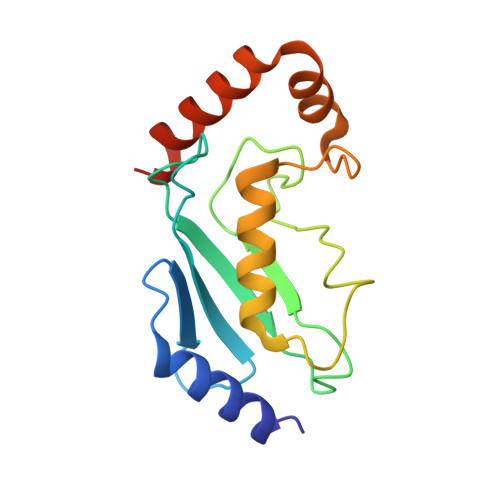
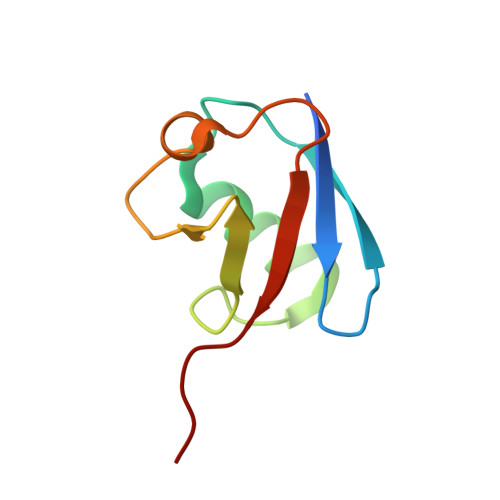
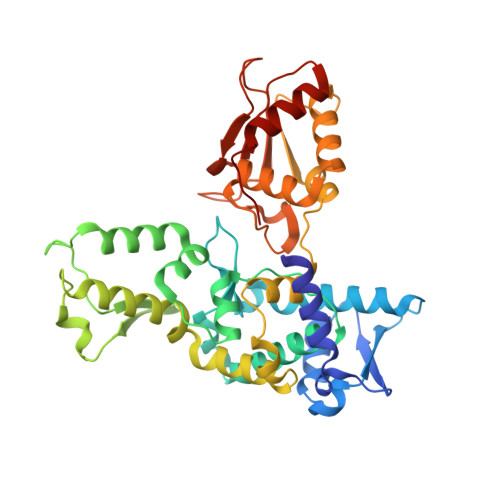
Structural basis for transthiolation intermediates in the ubiquitin pathway.
Kochanczyk, T., Hann, Z.S., Lux, M.C., Delos Reyes, A.M.V., Ji, C., Tan, D.S., Lima, C.D.(2024) Nature 633: 216-223
- PubMed: 39143218
- DOI: https://doi.org/10.1038/s41586-024-07828-9
- Primary Citation of Related Structures:
9B55, 9B56, 9B57, 9B58, 9B59, 9B5A, 9B5B, 9B5C, 9B5D, 9B5E, 9B5F, 9B5G, 9B5H, 9B5I, 9B5J, 9B5K, 9B5L, 9B5M, 9B5N, 9B5O, 9B5P, 9B5Q, 9B5R, 9B5S, 9B5T, 9B5U, 9B5V, 9B5W, 9B5X - PubMed Abstract:
Transthiolation (also known as transthioesterification) reactions are used in the biosynthesis of acetyl coenzyme A, fatty acids and polyketides, and for post-translational modification by ubiquitin (Ub) and ubiquitin-like (Ubl) proteins 1-3 . For the Ub pathway, E1 enzymes catalyse transthiolation from an E1~Ub thioester to an E2~Ub thioester. Transthiolation is also required for transfer of Ub from an E2~Ub thioester to HECT (homologous to E6AP C terminus) and RBR (ring-between-ring) E3 ligases to form E3~Ub thioesters 4-6 . How isoenergetic transfer of thioester bonds is driven forward by enzymes in the Ub pathway remains unclear. Here we isolate mimics of transient transthiolation intermediates for E1-Ub(T)-E2 and E2-Ub(T)-E3 HECT complexes (where T denotes Ub in a thioester or Ub undergoing transthiolation) using a chemical strategy with native enzymes and near-native Ub to capture and visualize a continuum of structures determined by single-particle cryo-electron microscopy. These structures and accompanying biochemical experiments illuminate conformational changes in Ub, E1, E2 and E3 that are coordinated with the chemical reactions to facilitate directional transfer of Ub from each enzyme to the next.
- Structural Biology Program, Sloan Kettering Institute, New York, NY, USA.
Organizational Affiliation: