A potential role for RNA aminoacylation prior to its role in peptide synthesis.
Radakovic, A., Lewicka, A., Todisco, M., Aitken, H.R.M., Weiss, Z., Kim, S., Bannan, A., Piccirilli, J.A., Szostak, J.W.(2024) Proc Natl Acad Sci U S A 121: e2410206121-e2410206121
- PubMed: 39178230
- DOI: https://doi.org/10.1073/pnas.2410206121
- Primary Citation of Related Structures:
9AUR, 9AUS - PubMed Abstract:
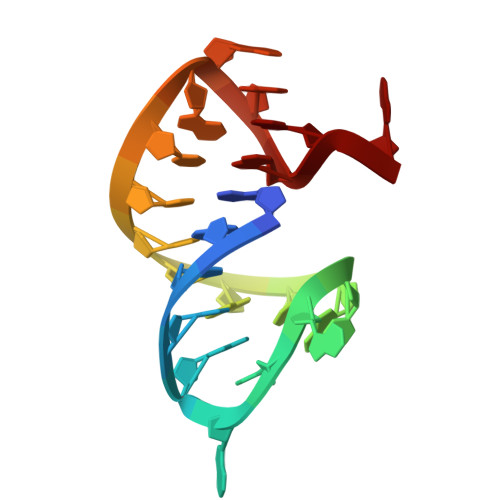
Coded ribosomal peptide synthesis could not have evolved unless its sequence and amino acid-specific aminoacylated tRNA substrates already existed. We therefore wondered whether aminoacylated RNAs might have served some primordial function prior to their role in protein synthesis. Here, we show that specific RNA sequences can be nonenzymatically aminoacylated and ligated to produce amino acid-bridged stem-loop RNAs. We used deep sequencing to identify RNAs that undergo highly efficient glycine aminoacylation followed by loop-closing ligation. The crystal structure of one such glycine-bridged RNA hairpin reveals a compact internally stabilized structure with the same eponymous T-loop architecture that is found in many noncoding RNAs, including the modern tRNA. We demonstrate that the T-loop-assisted amino acid bridging of RNA oligonucleotides enables the rapid template-free assembly of a chimeric version of an aminoacyl-RNA synthetase ribozyme. We suggest that the primordial assembly of amino acid-bridged chimeric ribozymes provides a direct and facile route for the covalent incorporation of amino acids into RNA. A greater functionality of covalently incorporated amino acids could contribute to enhanced ribozyme catalysis, providing a driving force for the evolution of sequence and amino acid-specific aminoacyl-RNA synthetase ribozymes in the RNA World. The synthesis of specifically aminoacylated RNAs, an unlikely prospect for nonenzymatic reactions but a likely one for ribozymes, could have set the stage for the subsequent evolution of coded protein synthesis.
- HHMI, Department of Chemistry, The University of Chicago, Chicago, IL 60637.
Organizational Affiliation: