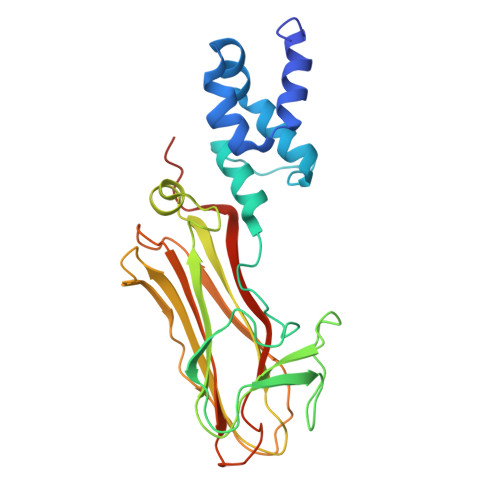
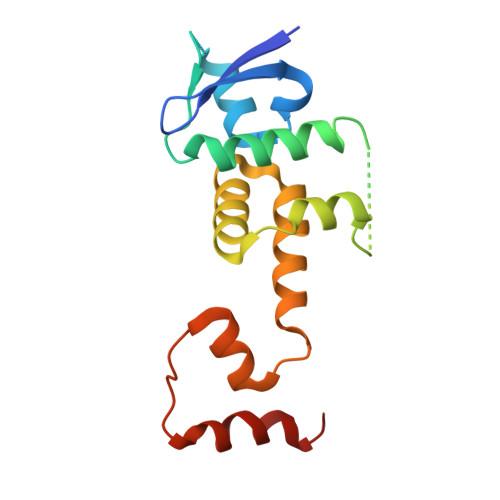
Structural basis of sugar recognition by SCF FBS2 ubiquitin ligase involved in NGLY1 deficiency.
Satoh, T., Yagi-Utsumi, M., Ishii, N., Mizushima, T., Yagi, H., Kato, R., Tachida, Y., Tateno, H., Matsuo, I., Kato, K., Suzuki, T., Yoshida, Y.(2024) FEBS Lett 598: 2259-2268
- PubMed: 39171510
- DOI: https://doi.org/10.1002/1873-3468.15003
- Primary Citation of Related Structures:
8ZUH - PubMed Abstract:
The cytosolic peptide:N-glycanase (PNGase) is involved in the quality control of N-glycoproteins via the endoplasmic reticulum-associated degradation (ERAD) pathway. Mutations in the gene encoding cytosolic PNGase (NGLY1 in humans) cause NGLY1 deficiency. Recent findings indicate that the F-box protein FBS2 of the SCF FBS2 ubiquitin ligase complex can be a promising drug target for NGLY1 deficiency. Here, we determined the crystal structure of bovine FBS2 complexed with the adaptor protein SKP1 and a sugar ligand, Man 3 GlcNAc 2 , which corresponds to the core pentasaccharide of N-glycan. Our crystallographic data together with NMR data revealed the structural basis of disparate sugar-binding specificities in homologous FBS proteins and identified a potential druggable pocket for in silico docking studies. Our results provide a potential basis for the development of selective inhibitors against FBS2 in NGLY1 deficiency.
- Graduate School of Pharmaceutical Sciences, Nagoya City University, Japan.
Organizational Affiliation: