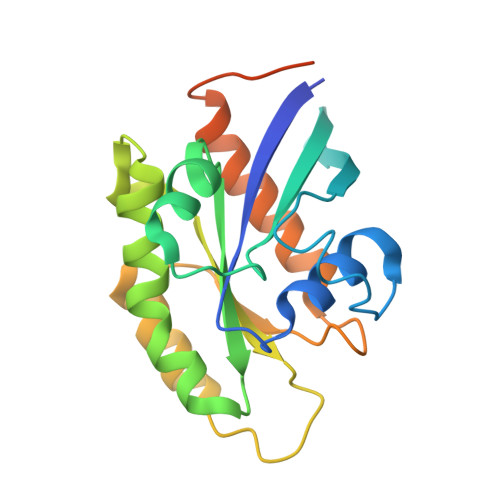
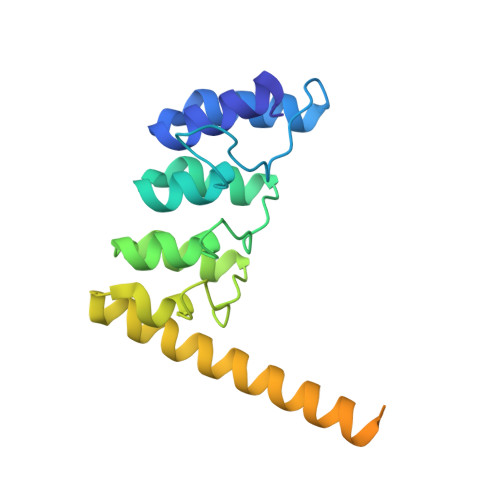
Structure of GDP-bound Rab7 Q67L in complex with ORP1L.
Lu, Q., Zhu, Z., Zhang, J., Xu, C.(2024) Biochem Biophys Res Commun 725: 150232-150232
- PubMed: 38897042
- DOI: https://doi.org/10.1016/j.bbrc.2024.150232
- Primary Citation of Related Structures:
8ZQ3 - PubMed Abstract:
Molecular processes are orchestrated by various proteins that promote early endosomes to become late endosomes and eventually fuse with lysosomes, guaranteeing the degradation of the content. Rab7, which is localized to late endosomes, is one of the most well-known GTPases. ORP1L is recruited by Rab7 to facilitate the fusion of late endosomes and lysosomes. Here, we present the structure of GDP-bound Rab7 Q67L with ORP1L. Structural analysis, supported by biochemical and ITC binding experiments, not only provides structural insight into the interactions between the ORP1L ANK domain and Rab7 but also suggests that the GTPase activity of Rab7 does not interfere with its ORP1L-binding capacity.
Organizational Affiliation:
Center for Advanced Interdisciplinary Science and Biomedicine of IHM, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, 230027, PR China.