Structure of the canthaxanthin mutant PSI-4VCPI supercomplex in Nannochloropsis oceanica
Shen, L.L., Li, Z.H., Shen, J.R., Wang, W.D.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| VCPI-9 | A [auth 9] | 232 | Nannochloropsis oceanica strain IMET1 | Mutation(s): 0 |  |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| VCPI-8 | B [auth 8] | 200 | Nannochloropsis oceanica strain IMET1 | Mutation(s): 0 |  |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| VCPI-7 | C [auth 7] | 202 | Nannochloropsis oceanica strain IMET1 | Mutation(s): 0 |  |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| VCPI-1 | D [auth 1] | 208 | Nannochloropsis oceanica strain IMET1 | Mutation(s): 0 |  |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem I P700 chlorophyll a apoprotein A1 | E [auth a] | 745 | Nannochloropsis oceanica strain IMET1 | Mutation(s): 0 EC: 1.97.1.12 | 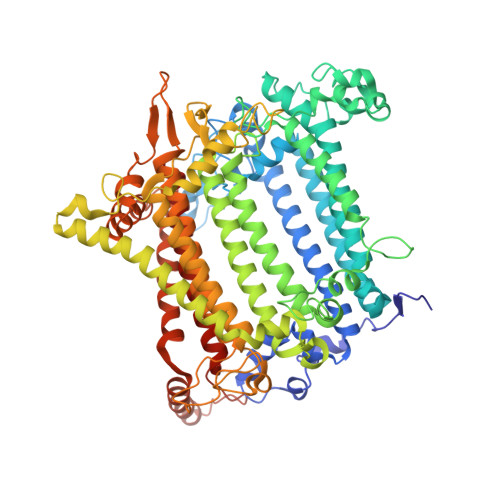 |
UniProt | |||||
Find proteins for T1RJP7 (Nannochloropsis oceanica) Explore T1RJP7 Go to UniProtKB: T1RJP7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | T1RJP7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem I P700 chlorophyll a apoprotein A2 | F [auth b] | 737 | Nannochloropsis oceanica strain IMET1 | Mutation(s): 0 EC: 1.97.1.12 | 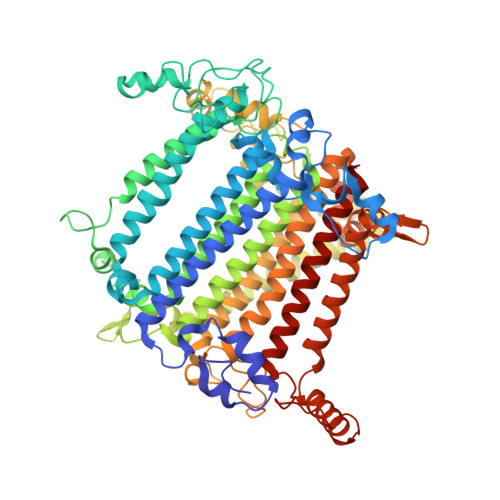 |
UniProt | |||||
Find proteins for T1RIQ1 (Nannochloropsis oceanica) Explore T1RIQ1 Go to UniProtKB: T1RIQ1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | T1RIQ1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem I reaction center subunit II | G [auth d] | 136 | Nannochloropsis oceanica strain IMET1 | Mutation(s): 0 | 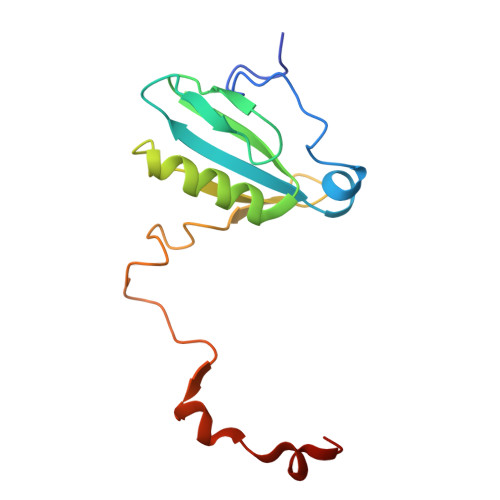 |
UniProt | |||||
Find proteins for T1RIT2 (Nannochloropsis oceanica) Explore T1RIT2 Go to UniProtKB: T1RIT2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | T1RIT2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 8 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem I reaction center subunit IV | H [auth e] | 67 | Nannochloropsis oceanica strain IMET1 | Mutation(s): 0 | 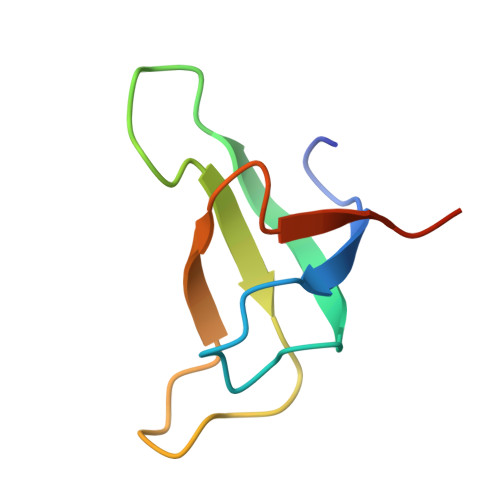 |
UniProt | |||||
Find proteins for T1RJT6 (Nannochloropsis oceanica) Explore T1RJT6 Go to UniProtKB: T1RJT6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | T1RJT6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 9 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem I reaction center subunit III | I [auth f] | 185 | Nannochloropsis oceanica strain IMET1 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for T1RJ05 (Nannochloropsis oceanica) Explore T1RJ05 Go to UniProtKB: T1RJ05 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | T1RJ05 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 10 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| PsaR | J [auth h] | 128 | Nannochloropsis oceanica strain IMET1 | Mutation(s): 0 |  |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 11 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem I reaction center subunit VIII | K [auth i] | 45 | Nannochloropsis oceanica strain IMET1 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for T1RJ07 (Nannochloropsis oceanica) Explore T1RJ07 Go to UniProtKB: T1RJ07 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | T1RJ07 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 12 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem I reaction center subunit IX | L [auth j] | 41 | Nannochloropsis oceanica strain IMET1 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for T1RJW8 (Nannochloropsis oceanica) Explore T1RJW8 Go to UniProtKB: T1RJW8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | T1RJW8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 13 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| PSI subunit V | M [auth l] | 172 | Nannochloropsis oceanica strain IMET1 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for T1RJZ1 (Nannochloropsis oceanica) Explore T1RJZ1 Go to UniProtKB: T1RJZ1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | T1RJZ1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 14 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| PsaM | N [auth m] | 30 | Nannochloropsis oceanica strain IMET1 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8F5PPL2 (Chaetoceros tenuissimus) Explore A0A8F5PPL2 Go to UniProtKB: A0A8F5PPL2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8F5PPL2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 15 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| PsaS | O [auth g] | 55 | Nannochloropsis oceanica strain IMET1 | Mutation(s): 0 |  |
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 16 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem I iron-sulfur center | P [auth c] | 81 | Nannochloropsis oceanica strain IMET1 | Mutation(s): 0 EC: 1.97.1.12 |  |
UniProt | |||||
Find proteins for T1RJY1 (Nannochloropsis oceanica) Explore T1RJY1 Go to UniProtKB: T1RJY1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | T1RJY1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 12 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| DGD (Subject of Investigation/LOI) Query on DGD | AG [auth b], WA [auth 8] | DIGALACTOSYL DIACYL GLYCEROL (DGDG) C51 H96 O15 LDQFLSUQYHBXSX-HXXRYREZSA-N |  | ||
| CLA (Subject of Investigation/LOI) Query on CLA | AA [auth 9] AC [auth 1] AD [auth a] AF [auth b] BA [auth 9] | CHLOROPHYLL A C55 H72 Mg N4 O5 ATNHDLDRLWWWCB-AENOIHSZSA-M |  | ||
| SQD (Subject of Investigation/LOI) Query on SQD | CC [auth 1] | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL C41 H78 O12 S RVUUQPKXGDTQPG-JUDHQOGESA-N |  | ||
| LMG (Subject of Investigation/LOI) Query on LMG | DE [auth a], QG [auth j] | 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE C45 H86 O10 DCLTVZLYPPIIID-CVELTQQQSA-N |  | ||
| A1L1F (Subject of Investigation/LOI) Query on A1L1F | JG [auth h], LA [auth 8], R [auth 9], RB [auth 1] | [(2~{Z},4~{E},6~{E},8~{E},10~{E},12~{E},14~{E})-17-[(4~{S},6~{R})-4-acetyloxy-2,2,6-trimethyl-6-oxidanyl-cyclohexylidene]-6,11,15-trimethyl-2-[(~{E})-2-[(1~{S},4~{S},6~{R})-2,2,6-trimethyl-4-oxidanyl-7-oxabicyclo[4.1.0]heptan-1-yl]ethenyl]heptadeca-2,4,6,8,10,12,14,16-octaenyl] octanoate C50 H72 O7 JYJFYJLLQQTZBJ-AYSGSIDZSA-N |  | ||
| LHG (Subject of Investigation/LOI) Query on LHG | GA [auth 9], VD [auth a], W [auth 9], WD [auth a], ZF [auth b] | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE C38 H75 O10 P BIABMEZBCHDPBV-MPQUPPDSSA-N |  | ||
| A1L1G (Subject of Investigation/LOI) Query on A1L1G | OB [auth 1], Q [auth 9], V [auth 9], YA [auth 7] | (1~{R},3~{S})-6-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{Z},17~{E})-16-(hydroxymethyl)-3,7,12-trimethyl-18-[(1~{S},4~{S},6~{R})-2,2,6-trimethyl-4-oxidanyl-7-oxabicyclo[4.1.0]heptan-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenylidene]-1,5,5-trimethyl-cyclohexane-1,3-diol C40 H56 O5 OWWSPDNLRSCPIS-XZHZJERWSA-N |  | ||
| XAT (Subject of Investigation/LOI) Query on XAT | AB [auth 7] BB [auth 7] CE [auth a] IA [auth 8] JA [auth 8] | (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL C40 H56 O4 SZCBXWMUOPQSOX-WVJDLNGLSA-N |  | ||
| 45D (Subject of Investigation/LOI) Query on 45D | U [auth 9] | beta,beta-carotene-4,4'-dione C40 H52 O2 FDSDTBUPSURDBL-DKLMTRRASA-N |  | ||
| BCR (Subject of Investigation/LOI) Query on BCR | AE [auth a] BG [auth b] CG [auth f] FG [auth f] GG [auth h] | BETA-CAROTENE C40 H56 OENHQHLEOONYIE-JLTXGRSLSA-N |  | ||
| PQN (Subject of Investigation/LOI) Query on PQN | TD [auth a], TF [auth b] | PHYLLOQUINONE C31 H46 O2 MBWXNTAXLNYFJB-NKFFZRIASA-N |  | ||
| SF4 (Subject of Investigation/LOI) Query on SF4 | BE [auth a], XG [auth c], YG [auth c] | IRON/SULFUR CLUSTER Fe4 S4 LJBDFODJNLIPKO-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Chinese Academy of Sciences | China | -- |