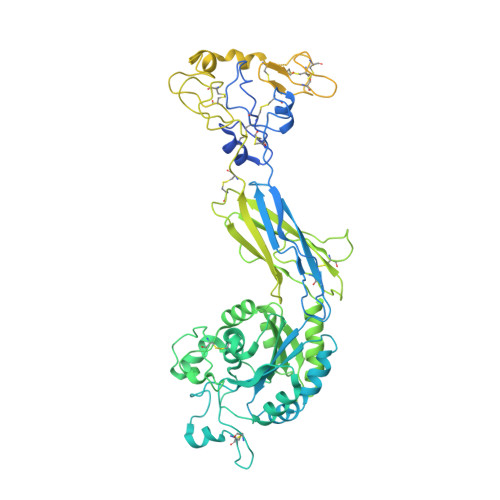
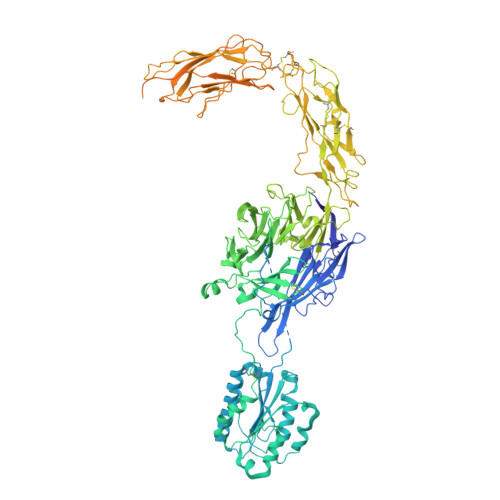Cryo-EM structure of I domain-containing integrin alpha E beta 7.
Akasaka, H., Sato, D., Shihoya, W., Nureki, O., Kise, Y.(2024) Biochem Biophys Res Commun 721: 150121-150121
- PubMed: 38781659
- DOI: https://doi.org/10.1016/j.bbrc.2024.150121
- Primary Citation of Related Structures:
8ZJF - PubMed Abstract:
The integrin family is a transmembrane receptor that plays critical roles in the cell-cell and cell-extracellular matrix adhesion, signal transduction such as cell cycle regulation, organization of the intracellular cytoskeleton, and immune responses. Consequently, dysfunction of integrins is associated with a wide range of human diseases, including cancer and immune diseases, which makes integrins therapeutic targets for drug discovery. Here we report the cryo-EM structure of the human α-I domain-containing full-length integrin αEβ7, which is expressed in the leukocytes of the immune system and a drug target for inflammatory bowel disease (IBD). The structure reveals the half-bent conformation, an intermediate between the close and the open conformation, while the α-I domain responsible for the ligand binding covers the headpiece domain by a unique spatial arrangement. Our results provide the structural information for the drug design targeting IBD.
- Department of Biological Sciences, Graduate School of Science, The University of Tokyo, Bunkyo, Tokyo, 113-0033, Japan.
Organizational Affiliation: