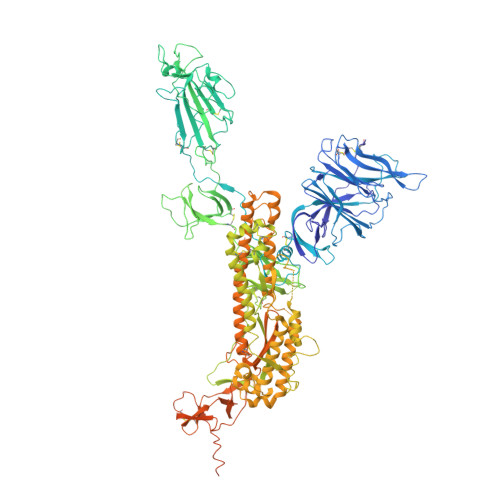
Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein and potentially drive the genesis of Omicron variants.
Yan, Q., Gao, X., Liu, B., Hou, R., He, P., Ma, Y., Zhang, Y., Zhang, Y., Li, Z., Chen, Q., Wang, J., Huang, X., Liang, H., Zheng, H., Yao, Y., Chen, X., Niu, X., He, J., Chen, L., Zhao, J., Xiong, X.(2024) Nat Commun 15: 7585-7585
- PubMed: 39217172
- DOI: https://doi.org/10.1038/s41467-024-51770-3
- Primary Citation of Related Structures:
8ZHD, 8ZHE, 8ZHF, 8ZHG, 8ZHH, 8ZHI, 8ZHJ, 8ZHK, 8ZHL, 8ZHM, 8ZHN, 8ZHO, 8ZHP - PubMed Abstract:
Continued evolution of SARS-CoV-2 generates variants to challenge antibody immunity established by infection and vaccination. A connection between population immunity and genesis of virus variants has long been suggested but its molecular basis remains poorly understood. Here, we identify a class of SARS-CoV-2 neutralizing public antibodies defined by their shared usage of VL6-57 light chains. Although heavy chains of diverse genotypes are utilized, convergent HCDR3 rearrangements have been observed among these public antibodies to cooperate with germline VL6-57 LCDRs to target a convergent epitope defined by RBD residues S371-S373-S375. Antibody repertoire analysis identifies that this class of VL6-57 antibodies is present in SARS-CoV-2-naive individuals and is clonally expanded in most COVID-19 patients. We confirm that Omicron-specific substitutions at S371, S373 and S375 mediate escape of antibodies of the VL6-57 class. These findings support that this class of public antibodies constitutes a potential immune pressure promoting the introduction of S371L/F-S373P-S375F in Omicron variants. The results provide further molecular evidence to support that antigenic evolution of SARS-CoV-2 is driven by antibody mediated population immunity.
- State Key Laboratory of Respiratory Disease, National Clinical Research Center for Respiratory Disease, Guangzhou Institute of Respiratory Health, the First Affiliated Hospital of Guangzhou Medical University, Guangzhou, China.
Organizational Affiliation: