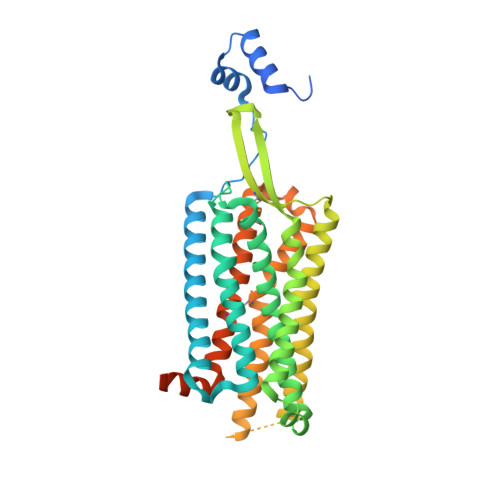
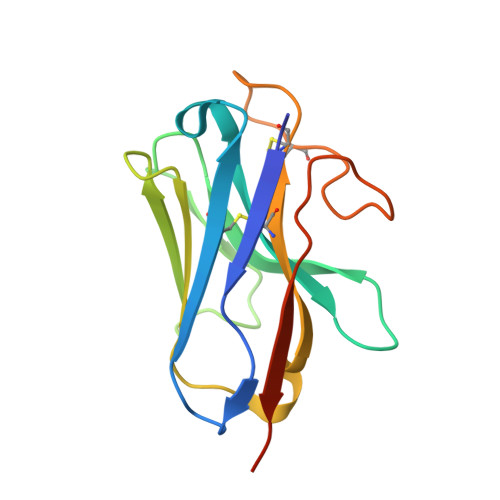
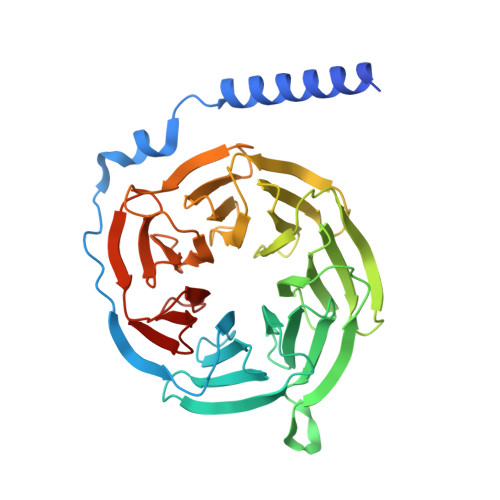
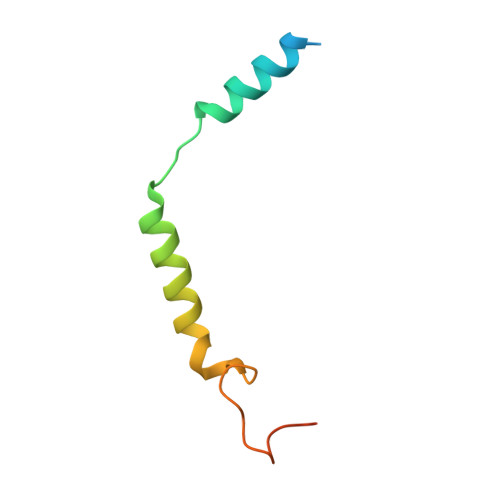
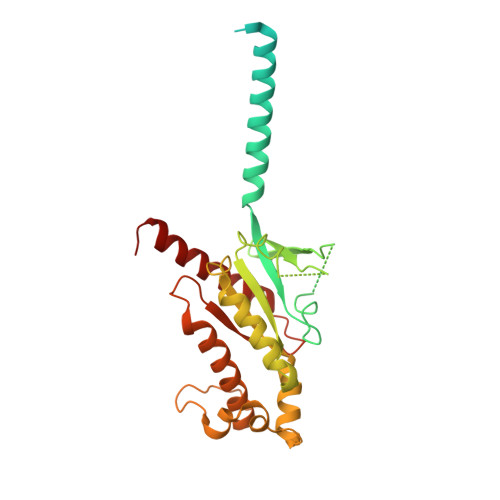
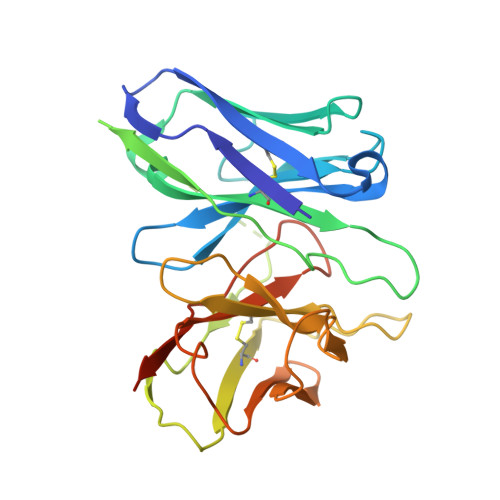
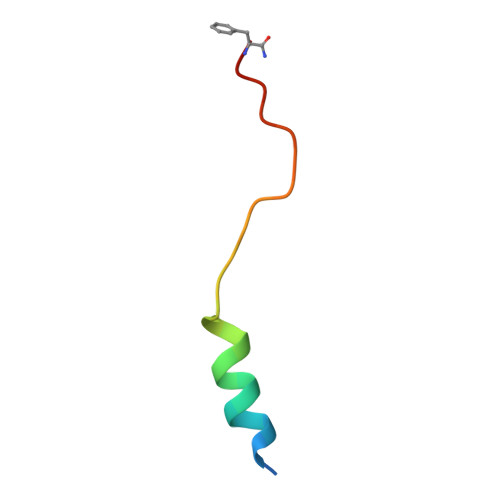
Structure and dynamics of the pyroglutamylated RF-amide peptide QRFP receptor GPR103.
Iwama, A., Kise, R., Akasaka, H., Sano, F.K., Oshima, H.S., Inoue, A., Shihoya, W., Nureki, O.(2024) Nat Commun 15: 4769-4769
- PubMed: 38897996
- DOI: https://doi.org/10.1038/s41467-024-49030-5
- Primary Citation of Related Structures:
8ZH8 - PubMed Abstract:
Pyroglutamylated RF-amide peptide (QRFP) is a peptide hormone with a C-terminal RF-amide motif. QRFP selectively activates a class A G-protein-coupled receptor (GPCR) GPR103 to exert various physiological functions such as energy metabolism and appetite regulation. Here, we report the cryo-electron microscopy structure of the QRFP26-GPR103-G q complex at 3.19 Å resolution. QRFP26 adopts an extended structure bearing no secondary structure, with its N-terminal and C-terminal sides recognized by extracellular and transmembrane domains of GPR103 respectively. This movement, reminiscent of class B1 GPCRs except for orientation and structure of the ligand, is critical for the high-affinity binding and receptor specificity of QRFP26. Mutagenesis experiments validate the functional importance of the binding mode of QRFP26 by GPR103. Structural comparisons with closely related receptors, including RY-amide peptide-recognizing GPCRs, revealed conserved and diversified peptide recognition mechanisms, providing profound insights into the biological significance of RF-amide peptides. Collectively, this study not only advances our understanding of GPCR-ligand interactions, but also paves the way for the development of novel therapeutics targeting metabolic and appetite disorders and emergency medical care.
- Department of Biological Sciences, Graduate School of Science, The University of Tokyo, Bunkyo, Tokyo, 113-0033, Japan.
Organizational Affiliation: