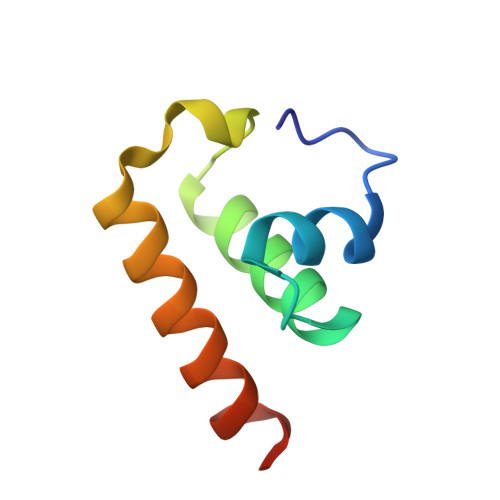
Novel structure of the anti-CRISPR protein AcrIE3 and its implication on the CRISPR-Cas inhibition.
Kim, D.Y., Han, J.H., Lee, S.Y., Ha, H.J., Park, H.H.(2024) Biochem Biophys Res Commun 722: 150164-150164
- PubMed: 38797150
- DOI: https://doi.org/10.1016/j.bbrc.2024.150164
- Primary Citation of Related Structures:
8ZEY - PubMed Abstract:
As a response to viral infections, bacteria have evolved the CRISPR-Cas system as an adaptive immune mechanism, enabling them to target and eliminate viral genetic material introduced during infection. However, viruses have also evolved mechanisms to counteract this bacterial defense, including anti-CRISPR proteins, which can inactivate the CRISPR-Cas adaptive immune system, thus aiding the viruses in their survival and replication within bacterial hosts. In this study, we establish the high-resolution crystal structure of the Type IE anti-CRISPR protein, AcrIE3. Our structural examination showed that AcrIE3 adopts a helical bundle fold comprising four α-helices, with a notably extended loop at the N-terminus. Additionally, surface analysis of AcrIE3 revealed the presence of three acidic regions, which potentially play a crucial role in the inhibitory function of this protein. The structural information we have elucidated for AcrIE3 will provide crucial insights into fully understanding its inhibitory mechanism. Furthermore, this information is anticipated to be important for the application of the AcrIE family in genetic editing, paving the way for advancements in gene editing technologies.
Organizational Affiliation:
College of Pharmacy, Chung-Ang University, Seoul, 06974, Republic of Korea; Department of Global Innovative Drugs, Graduate School of Chung-Ang University, Seoul, 06974, Republic of Korea.