Mechanism of Intracellular Elemental Sulfur Oxidation in Beggiatoa leptomitoformis , Where Persulfide Dioxygenase Plays a Key Role.
Rudenko, T.S., Trubitsina, L.I., Terentyev, V.V., Trubitsin, I.V., Borshchevskiy, V.I., Tishchenko, S.V., Gabdulkhakov, A.G., Leontievsky, A.A., Grabovich, M.Y.(2024) Int J Mol Sci 25
- PubMed: 39456744
- DOI: https://doi.org/10.3390/ijms252010962
- Primary Citation of Related Structures:
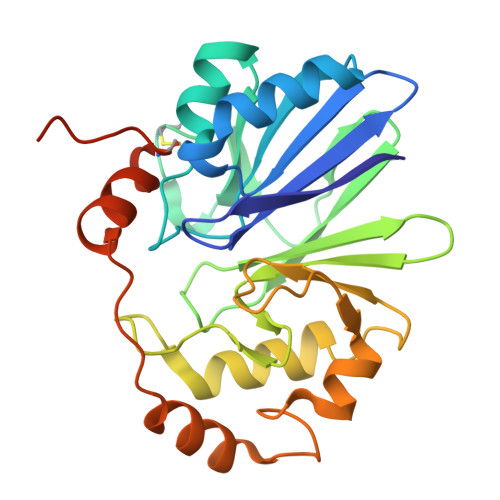
8ZBD - PubMed Abstract:
Representatives of the colorless sulfur bacteria of the genus Beggiatoa use reduced sulfur compounds in the processes of lithotrophic growth, which is accompanied by the storage of intracellular sulfur. However, it is still unknown how the transformation of intracellular sulfur occurs in Beggiatoa representatives. Annotation of the genome of Beggiatoa leptomitoformis D-402 did not identify any genes for the oxidation or reduction of elemental sulfur. By searching BLASTP, two putative persulfide dioxygenase (PDO) homologs were found in the genome of B. leptomitoformis . In some heterotrophic prokaryotes, PDO is involved in the oxidation of sulfane sulfur. According to HPLC-MS/MS, the revealed protein was reliably detected in a culture sample grown only in the presence of endogenous sulfur and CO 2 . The recombinant protein from B. leptomitoformis was active in the presence of glutathione persulfide. The crystal structure of recombinant PDO exhibited consistency with known structures of type I PDO. Thus, it was shown that B. leptomitoformis uses PDO to oxidize endogenous sulfur. Additionally, on the basis of HPLC-MS/MS, RT-qPCR, and the study of PDO reaction products, we predicted the interrelation of PDO and Sox-system function in the oxidation of endogenous sulfur in B. leptomitoformis and the connection of this process with energy metabolism.
- Department of Biochemistry and Cell Physiology, Voronezh State University, 394018 Voronezh, Russia.
Organizational Affiliation: