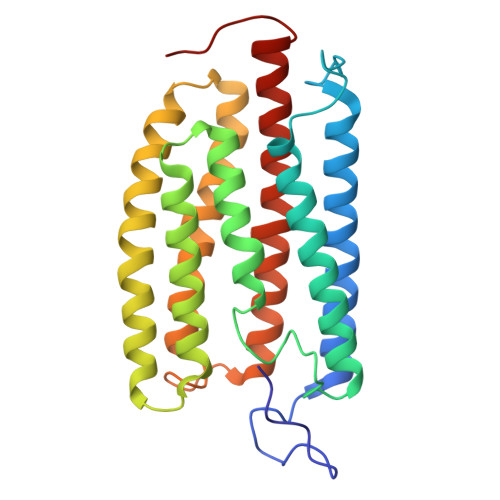
Channelrhodopsins with distinct chromophores and binding patterns.
Shan, Y., Zhao, L., Chen, M., Li, X., Zhang, M., Pei, D.(2024) Nat Commun 15: 7292
- PubMed: 39181878
- DOI: https://doi.org/10.1038/s41467-024-51811-x
- Primary Citation of Related Structures:
8ZAM, 8ZAN, 8ZAO, 8ZAP, 8ZAQ - PubMed Abstract:
Channelrhodopsins are popular optogenetic tools in neuroscience, but remain poorly understood mechanistically. Here we report the cryo-EM structures of channelrhodopsin-2 (ChR2) from Chlamydomonas reinhardtii and H. catenoides kalium channelrhodopsin (KCR1). We show that ChR2 recruits an endogenous N-retinylidene-PE-like molecule to a previously unidentified lateral retinal binding pocket, exhibiting a reduced light response in HEK293 cells. In contrast, H. catenoides kalium channelrhodopsin (KCR1) binds an endogenous retinal in its canonical retinal binding pocket under identical condition. However, exogenous ATR reduces the photocurrent magnitude of wild type KCR1 and also inhibits its leaky mutant C110T. Our results uncover diverse retinal chromophores with distinct binding patterns for channelrhodopsins in mammalian cells, which may further inspire next generation optogenetics for complex tasks such as cell fate control.
- Laboratory of Cell Fate Control, School of Life Sciences, Westlake University, Hangzhou, China.
Organizational Affiliation: