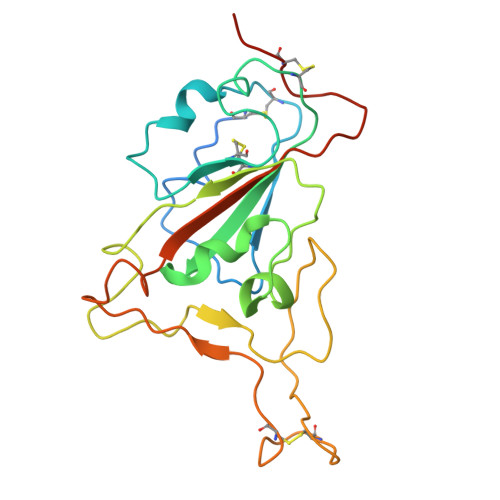
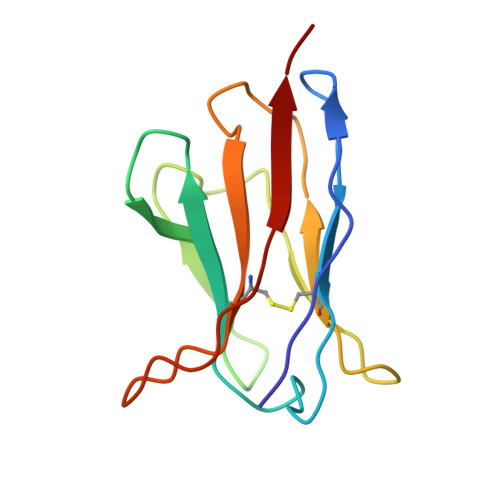
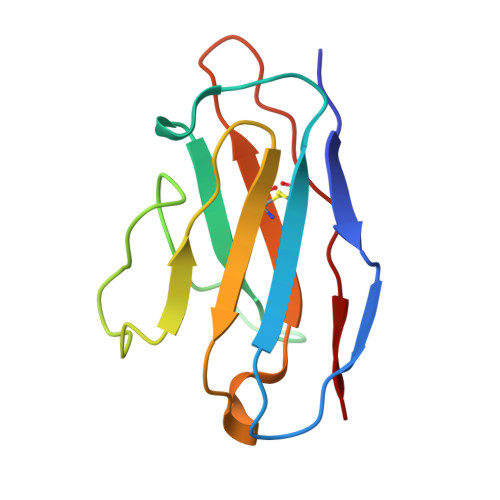
A broadly neutralizing antibody against the SARS-CoV-2 Omicron sub-variants BA.1, BA.2, BA.2.12.1, BA.4, and BA.5.
Chen, Z., Feng, L., Wang, L., Zhang, L., Zheng, B., Fu, H., Li, F., Liu, L., Lv, Q., Deng, R., Xu, Y., Hu, Y., Zheng, J., Qin, C., Bao, L., Wang, X., Jin, Q.(2025) Signal Transduct Target Ther 10: 14-14
- PubMed: 39800731
- DOI: https://doi.org/10.1038/s41392-024-02114-6
- Primary Citation of Related Structures:
8XSD, 8Z86 - PubMed Abstract:
The global spread of Severe Acute Respiratory Syndrome Coronavirus 2. (SARS-CoV-2) and its variant strains, including Alpha, Beta, Gamma, Delta, and now Omicron, pose a significant challenge. With the constant evolution of the virus, Omicron and its subtypes BA.1, BA.2, BA.3, BA.4, and BA.5 have developed the capacity to evade neutralization induced by previous vaccination or infection. This evasion highlights the urgency in discovering new monoclonal antibodies (mAbs) with neutralizing activity, especially broadly neutralizing antibodies (bnAbs), to combat the virus.In the current study, we introduced a fully human neutralizing mAb, CR9, that targets Omicron variants. We demonstrated the mAb's effectiveness in inhibiting Omicron replication both in vitro and in vivo. Structural analysis using cryo-electron microscopy (cryo-EM) revealed that CR9 binds to an epitope formed by RBD residues, providing a molecular understanding of its neutralization mechanism. Given its potency and specificity, CR9 holds promise as a potential adjunct therapy for treating Omicron infections. Our findings highlight the importance of continuous mAb discovery and characterization in addressing the evolving threat of COVID-19.
- NHC Key Laboratory of Systems Biology of Pathogens, State Key Laboratory of Respiratory Health and Multimorbidity, National Institute of Pathogen Biology, and Center for Tuberculosis Research, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, 100730, China.
Organizational Affiliation: