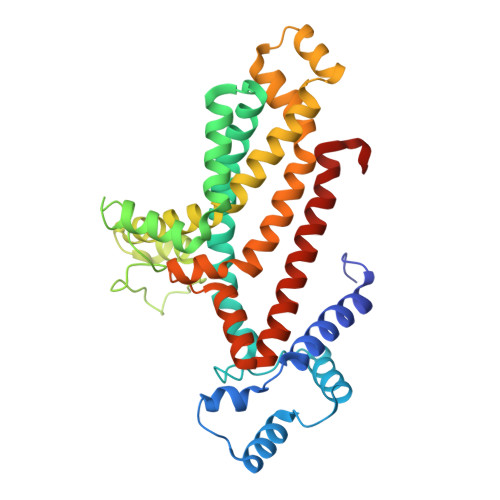
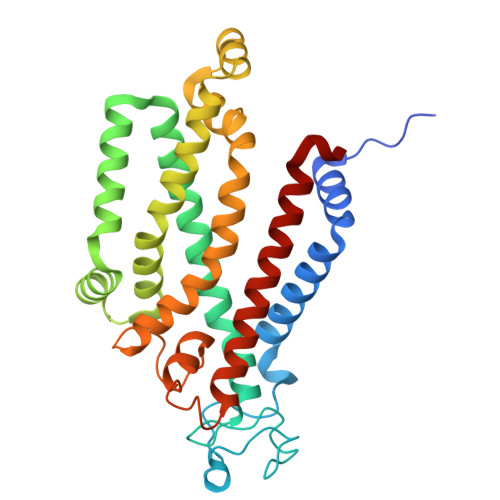
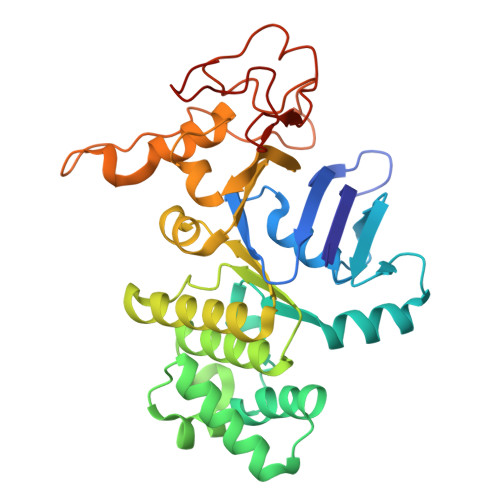
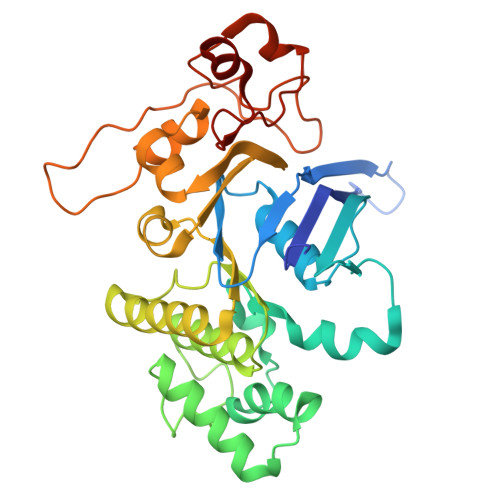
Structural characterization of the ABC transporter DppABCDF in Escherichia coli reveals insights into dipeptide acquisition.
Li, P., Zhang, M., Huang, Y.(2025) PLoS Biol 23: e3003026-e3003026
- PubMed: 40053564
- DOI: https://doi.org/10.1371/journal.pbio.3003026
- Primary Citation of Related Structures:
8Z1V, 8Z1W, 8Z1X, 8Z1Y - PubMed Abstract:
The prokaryote-specific ATP-binding cassette (ABC) peptide transporters are involved in various physiological processes and plays an important role in transporting naturally occurring antibiotics across the membrane to their intracellular targets. The dipeptide transporter DppABCDF in Gram-negative bacteria is composed of five distinct subunits, yet its assembly and underlying peptide import mechanism remain elusive. Here, we report the cryo-EM structures of the DppBCDF translocator from Escherichia coli in both its apo form and in complexes bound to nonhydrolyzable or slowly hydrolyzable ATP analogs (AMPPNP and ATPγS), as well as the ATPγS-bound DppABCDF full transporter. Unlike the reported heterotrimeric Mycobacterium tuberculosis DppBCD translocator, the E. coli DppBCDF translocator is a heterotetramer, with a [4Fe-4S] cluster at the C-terminus of each ATPase subunit. Structural studies reveal that ATPγS/AMPPNP-bound DppBCDF adopts an inward-facing conformation, similar to that of apo-DppBCDF, with only one ATPγS or AMPPNP molecule bound to DppF. By contrast, ATPγS-bound DppABCDF adopts an outward-facing conformation, with two ATPγS molecules glueing DppD and DppF at the interface. Consistent with structural observations, ATPase activity assays show that the DppBCDF translocator itself is inactive and its activation requires concurrent binding of DppA and ATP. In addition, bacterial complementation experiments imply that a unique periplasmic scoop motif in DppB may play important roles in ensuring dipeptide substrates import across the membrane, presumably by preventing dipeptide back-and-forth binding to DppA and avoiding dipeptides escaping into the periplasm upon being released from DppA.
- National Laboratory of Biomacromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing, China.
Organizational Affiliation: