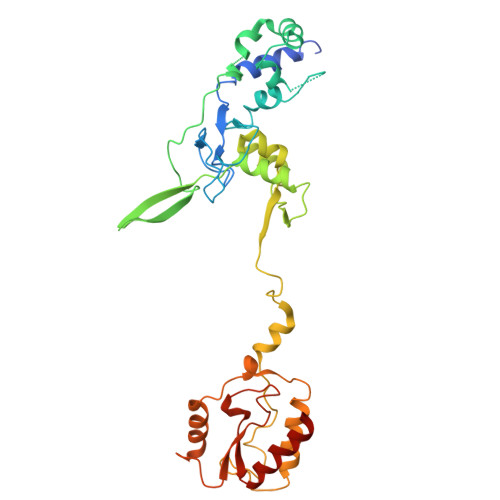
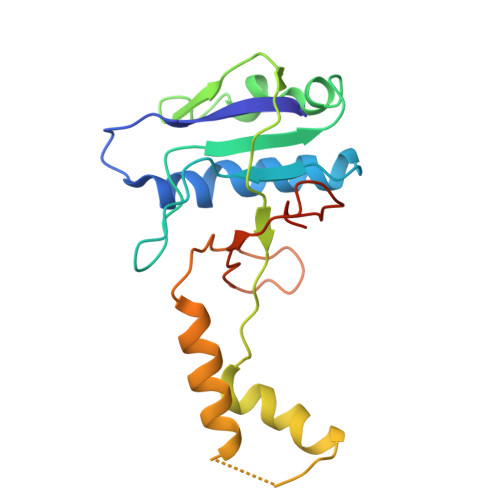
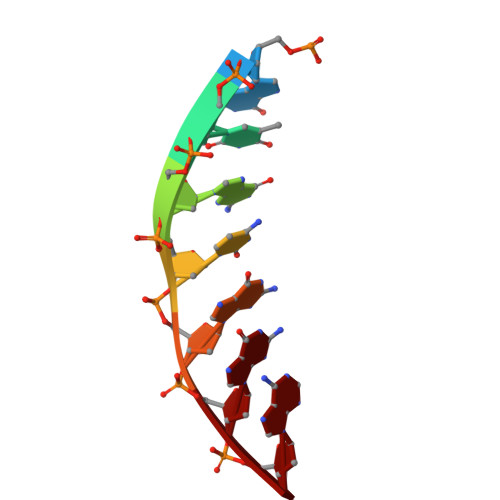
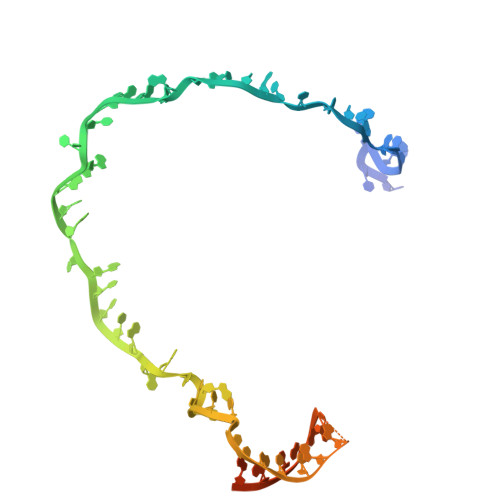
Structural basis for the type I-F Cas8-HNH system.
Li, X., Liu, Y., Han, J., Zhang, L., Liu, Z., Wang, L., Zhang, S., Zhang, Q., Fu, P., Yin, H., Zhu, H., Zhang, H.(2024) EMBO J 43: 4656-4667
- PubMed: 39251884
- DOI: https://doi.org/10.1038/s44318-024-00229-8
- Primary Citation of Related Structures:
8Z0K, 8Z0L, 8ZDY, 8ZNR - PubMed Abstract:
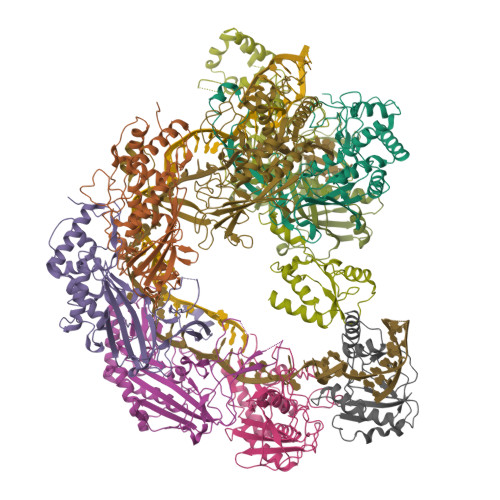
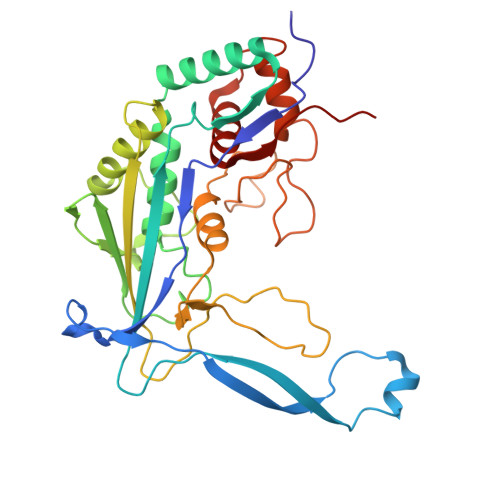
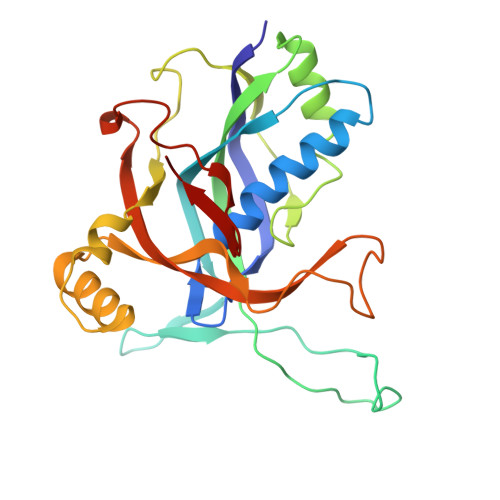
The Cas3 nuclease is utilized by canonical type I CRISPR-Cas systems for processive target DNA degradation, while a newly identified type I-F CRISPR variant employs an HNH nuclease domain from the natural fusion Cas8-HNH protein for precise target cleavage both in vitro and in human cells. Here, we report multiple cryo-electron microscopy structures of the type I-F Cas8-HNH system at different functional states. The Cas8-HNH Cascade complex adopts an overall G-shaped architecture, with the HNH domain occupying the C-terminal helical bundle domain (HB) of the Cas8 protein in canonical type I systems. The Linker region connecting Cas8-NTD and HNH domains adopts a rigid conformation and interacts with the Cas7.6 subunit, enabling the HNH domain to be in a functional position. The full R-loop formation displaces the HNH domain away from the Cas6 subunit, thus activating the target DNA cleavage. Importantly, our results demonstrate that precise target cleavage is dictated by a C-terminal helix of the HNH domain. Together, our work not only delineates the structural basis for target recognition and activation of the type I-F Cas8-HNH system, but also guides further developments leveraging this system for precise DNA editing.
Organizational Affiliation:
Tianjin Institute of Immunology, State Key Laboratory of Experimental Hematology, International Joint Laboratory of Ocular Diseases (Ministry of Education), Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), The Province and Ministry Co-sponsored Collaborative Innovation Center for Medical Epigenetics, School of Basic Medical Sciences, Tianjin Medical University, Tianjin, 300070, China.