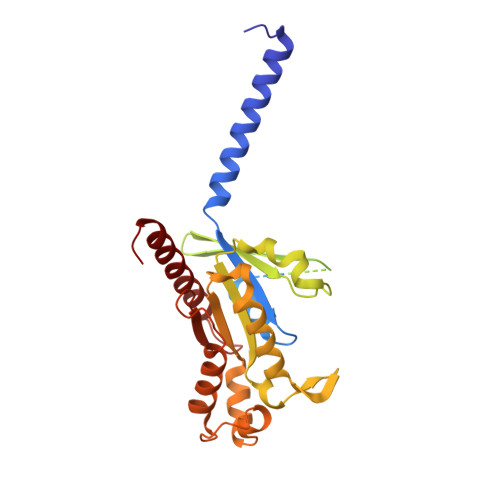
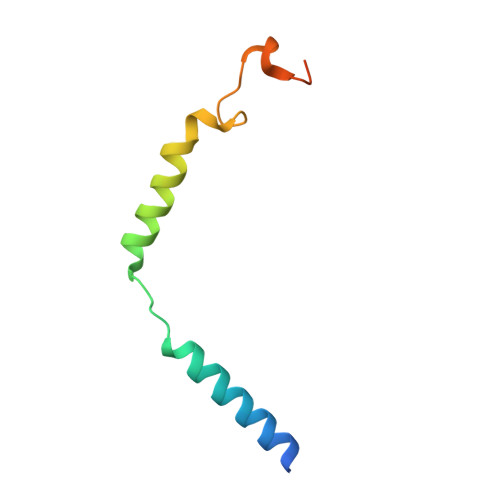
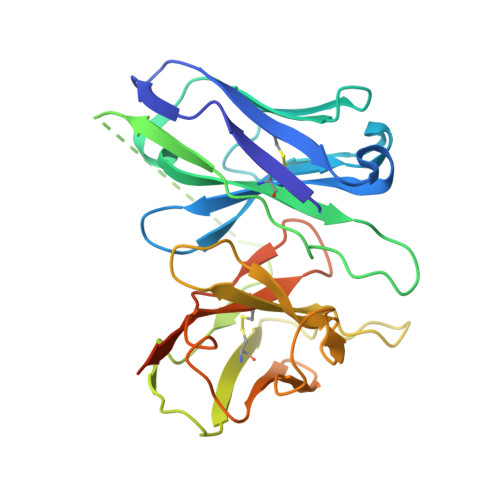
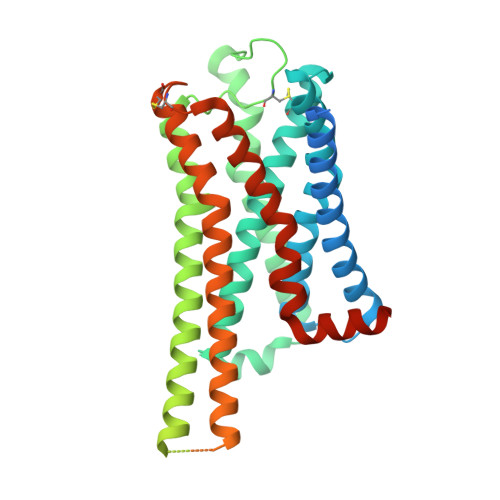
Molecular Determinant Underlying Selective Coupling of Primary G-Protein by Class A GPCRs.
Shen, Q., Tang, X., Wen, X., Cheng, S., Xiao, P., Zang, S.K., Shen, D.D., Jiang, L., Zheng, Y., Zhang, H., Xu, H., Mao, C., Zhang, M., Hu, W., Sun, J.P., Zhang, Y., Chen, Z.(2024) Adv Sci (Weinh) 11: e2310120-e2310120
- PubMed: 38647423
- DOI: https://doi.org/10.1002/advs.202310120
- Primary Citation of Related Structures:
8YUT, 8YUU, 8YUV - PubMed Abstract:
G-protein-coupled receptors (GPCRs) transmit downstream signals predominantly via G-protein pathways. However, the conformational basis of selective coupling of primary G-protein remains elusive. Histamine receptors H 2 R and H 3 R couple with G s - or G i -proteins respectively. Here, three cryo-EM structures of H 2 R-G s and H 3 R-G i complexes are presented at a global resolution of 2.6-2.7 Å. These structures reveal the unique binding pose for endogenous histamine in H 3 R, wherein the amino group interacts with E206 5.46 of H 3 R instead of the conserved D114 3.32 of other aminergic receptors. Furthermore, comparative analysis of the H 2 R-G s and H 3 R-G i complexes reveals that the structural geometry of TM5/TM6 determines the primary G-protein selectivity in histamine receptors. Machine learning (ML)-based structuromic profiling and functional analysis of class A GPCR-G-protein complexes illustrate that TM5 length, TM5 tilt, and TM6 outward movement are key determinants of the G s and G i/o selectivity among the whole Class A family. Collectively, the findings uncover the common structural geometry within class A GPCRs that determines the primary G s - and G i/o -coupling selectivity.
- Department of Pharmacology and Department of Pathology of Sir Run Run Shaw Hospital & Liangzhu Laboratory, Hangzhou, 310058, China.
Organizational Affiliation: