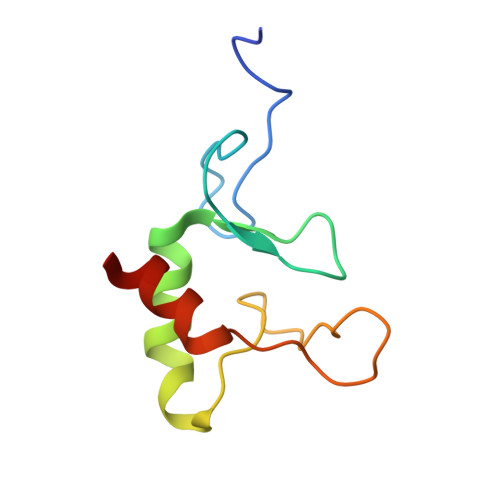
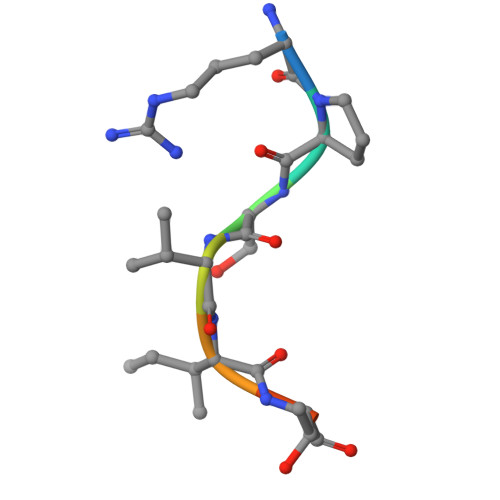
IRF2BP2 binds to a conserved RxSVI motif of protein partners and regulates megakaryocytic differentiation.
Wang, G., Lu, T., Zhang, L., Ding, J.(2024) Nat Commun 15: 10425-10425
- PubMed: 39616187
- DOI: https://doi.org/10.1038/s41467-024-54889-5
- Primary Citation of Related Structures:
8YTF, 8YTG, 8YTH - PubMed Abstract:
IRF2BP2 is a transcriptional coregulator that plays diverse regulatory roles in various cellular processes in either IRF2-dependent or IRF2-independent manner through interactions with protein partners via its RING domain; however, the underlying molecular mechanisms remain unclear. In this study, we conduct a motif discovery search on the sequences of interacting proteins IRF2 and VGLL4 of IRF2BP2 and identify a conserved RxSVI motif. Biochemical and structural data reveal that the RING domain binds to the motif-containing peptides of IRF2 and VGLL4 with comparable affinities and in a similar manner. The motif-containing peptides tend to form a short loop along with a short β-strand, which facilitates effective recognition and tight binding by the RING domain. Further exploration of this motif in the human proteome identifies the transcription factor ZBTB16 as an interacting protein of IRF2BP2. Biochemical, structural, and cell biological data demonstrate that the RING domain binds to the motif-containing peptide of ZBTB16 in a manner similar to that of IRF2 and VGLL4. Moreover, IRF2BP2 plays a crucial regulatory role in megakaryocytic differentiation through interaction with ZBTB16. These findings elucidate the molecular basis for how IRF2BP2 can engage with different protein partners, thereby exerting diverse regulatory functions in many cellular processes.
- Key Laboratory of RNA Innovation, Science and Engineering, Shanghai Institute of Biochemistry and Cell Biology, Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences, University of Chinese Academy of Sciences, Shanghai, China.
Organizational Affiliation: