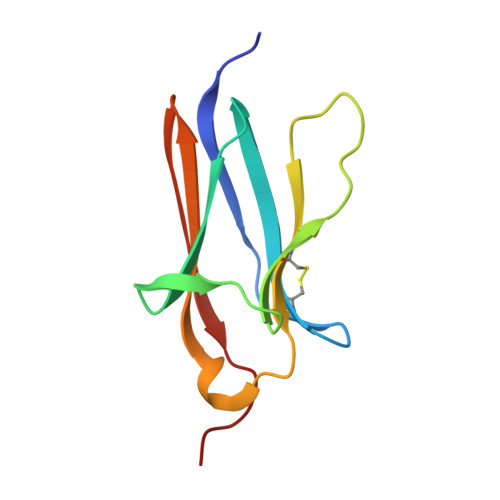
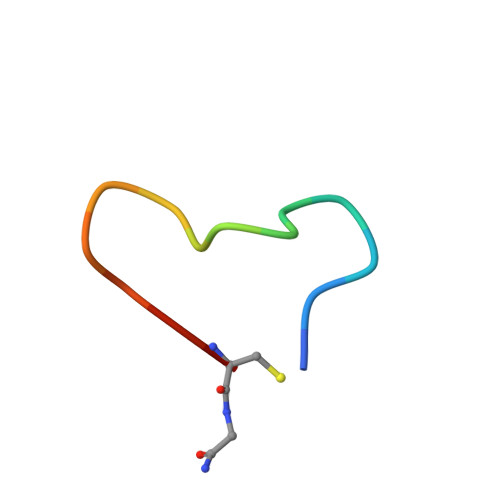
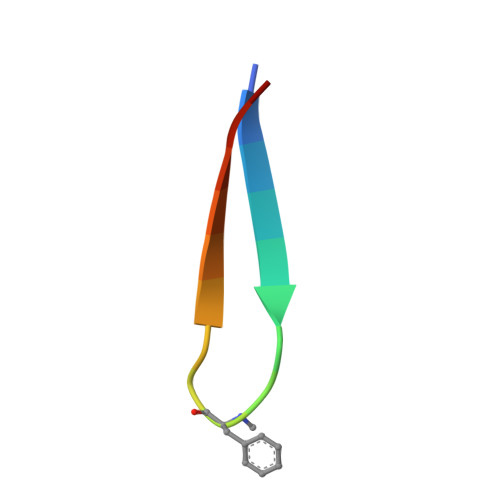
Discovery and Hit to Lead Optimization of Macrocyclic Peptides as Novel Tropomyosin Receptor Kinase A Antagonists.
Yamada, T., Mihara, K., Ueda, T., Yamauchi, D., Shimizu, M., Ando, A., Mayumi, K., Nakata, Z., Mikamiyama, H.(2024) J Med Chem 67: 11197-11208
- PubMed: 38950284
- DOI: https://doi.org/10.1021/acs.jmedchem.4c00715
- Primary Citation of Related Structures:
8YTD, 8YTE - PubMed Abstract:
Tropomyosin receptor kinases (Trks) are receptor tyrosine kinases activated by neurotrophic factors, called neurotrophins. Among them, TrkA interacts with the nerve growth factor (NGF), which leads to pain induction. mRNA-display screening was carried out to discover a hit compound 2 , which inhibits protein-protein interactions between TrkA and NGF. Subsequent structure optimization improving phosphorylation inhibitory activity and serum stability was pursued using a unique process that took advantage of the peptide being synthesized by translation from mRNA. This gave peptide 19 , which showed an analgesic effect in a rat incisional pain model. The peptides described here can serve as a new class of analgesics, and the structure optimization methods reported provide a strategy for discovering new peptide drugs.
- Biopharmaceutical Research Division, Shionogi Pharmaceutical Research Center, Toyonaka , Osaka 561-0825, Japan.
Organizational Affiliation: