Discovery of Novel LSD1 Inhibitors for the Treatment of Autosomal Dominant Polycystic Kidney Disease
Jiang, H., Tongchao, L., Bing, X., Zhiyan, D., Danyan, C.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Lysine-specific histone demethylase 1A | 660 | Homo sapiens | Mutation(s): 0 Gene Names: KDM1A, AOF2, KDM1, KIAA0601, LSD1 EC: 1.14.99.66 (PDB Primary Data), 1.14.11 (UniProt), 1.14.11.65 (UniProt) | 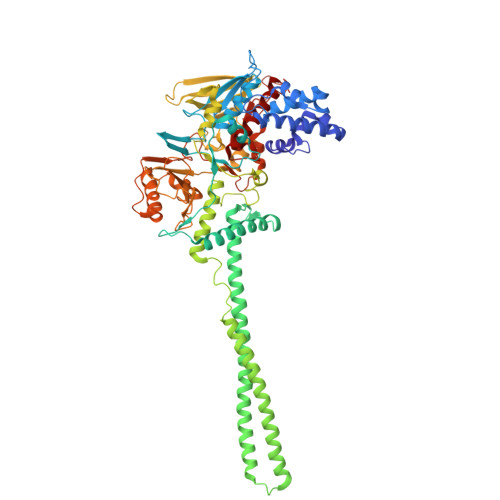 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for O60341 (Homo sapiens) Explore O60341 Go to UniProtKB: O60341 | |||||
PHAROS: O60341 GTEx: ENSG00000004487 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O60341 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| REST corepressor 1 | 134 | Homo sapiens | Mutation(s): 0 Gene Names: RCOR1, KIAA0071, RCOR | 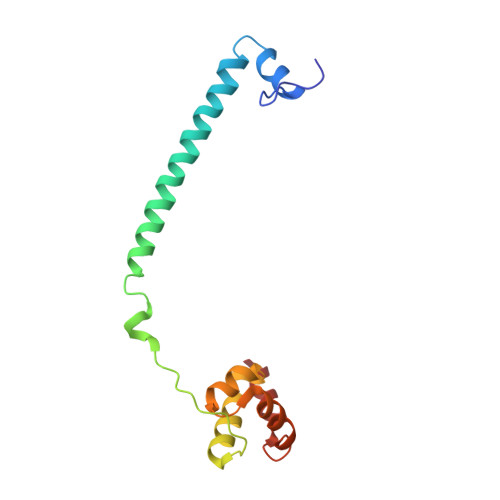 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9UKL0 (Homo sapiens) Explore Q9UKL0 Go to UniProtKB: Q9UKL0 | |||||
PHAROS: Q9UKL0 GTEx: ENSG00000089902 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9UKL0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| FAD (Subject of Investigation/LOI) Query on FAD | C [auth A] | FLAVIN-ADENINE DINUCLEOTIDE C27 H33 N9 O15 P2 VWWQXMAJTJZDQX-UYBVJOGSSA-N |  | ||
| A1LZJ (Subject of Investigation/LOI) Query on A1LZJ | D [auth A] | 4-[5-(4-azanylpiperidin-1-yl)-8-(4-methylphenyl)pyrido[3,4-b]pyrazin-7-yl]-2-fluoranyl-benzenecarbonitrile C26 H23 F N6 BISBXLFNGMDQKO-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 120.674 | α = 90 |
| b = 178.038 | β = 90 |
| c = 234.346 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Not funded | -- |