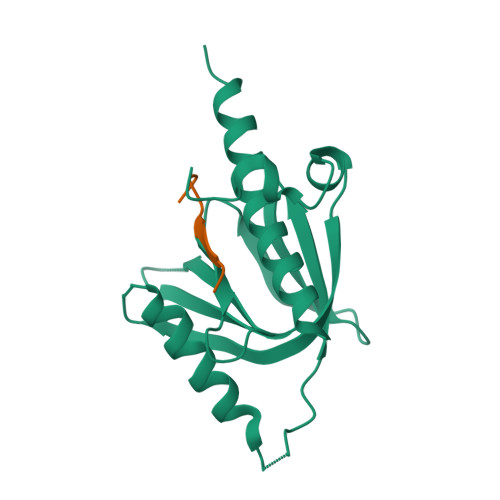
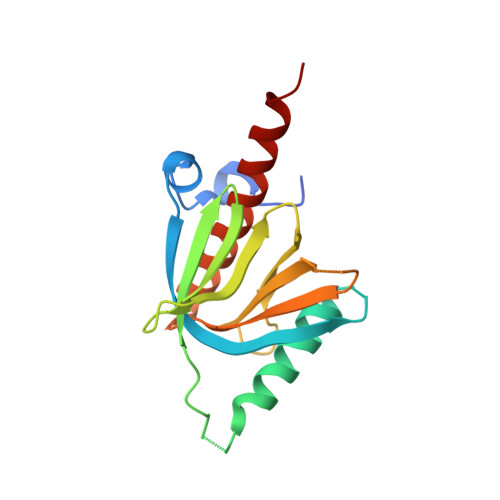
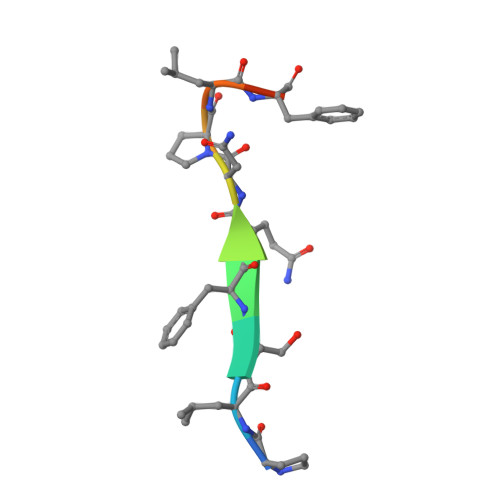
AIDA-1/ANKS1B Binds to the SynGAP Family RasGAPs with High Affinity and Specificity.
Wang, X., Wang, Y., Cai, Q., Zhang, M.(2024) J Mol Biology 436: 168608-168608
- PubMed: 38759928
- DOI: https://doi.org/10.1016/j.jmb.2024.168608
- Primary Citation of Related Structures:
8YM2 - PubMed Abstract:
AIDA-1, encoded by ANKS1B, is an abundant postsynaptic scaffold protein essential for brain development. Mutations of ANKS1B are closely associated with various psychiatric disorders. However, very little is known regarding the molecular mechanisms underlying AIDA-1's involvements under physiological and pathophysiological conditions. Here, we discovered an interaction between AIDA-1 and the SynGAP family Ras-GTPase activating protein (GAP) via affinity purification using AIDA-1d as the bait. Biochemical studies showed that the PTB domain of AIDA-1 binds to an extended NPx[F/Y]-motif of the SynGAP family proteins with high affinities. The high-resolution crystal structure of AIDA-1 PTB domain in complex with the SynGAP NPxF-motif revealed the molecular mechanism governing the specific interaction between AIDA-1 and SynGAP. Our study not only explains why patients with ANKS1B or SYNGAP1 mutations share overlapping clinical phenotypes, but also allows identification of new AIDA-1 binding targets such as Ras and Rab interactors.
Organizational Affiliation:
Biomedical Research Institute, Shenzhen Peking University-The Hong Kong University of Science and Technology Medical Center, Shenzhen 518036, China. Electronic address: xueqwang@pku.edu.cn.