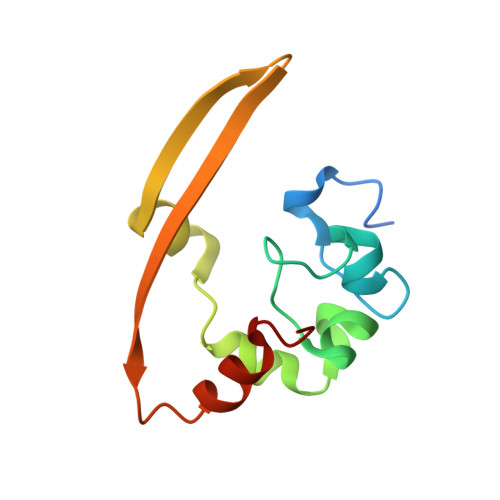
Unraveling the assembly mechanism of SADS-CoV virus nucleocapsid protein: insights from RNA binding, dimerization, and epitope diversity profiling.
Zhang, Y., Wu, F., Han, Y., Wu, Y., Huang, L., Huang, Y., Yan, D., Jiang, X., Ma, J., Xu, W.(2024) J Virol 98: e0092624-e0092624
- PubMed: 39082816
- DOI: https://doi.org/10.1128/jvi.00926-24
- Primary Citation of Related Structures:
8YM1 - PubMed Abstract:
The swine acute diarrhea syndrome coronavirus (SADS-CoV) has caused significant disruptions in porcine breeding and raised concerns about potential human infection. The nucleocapsid (N) protein of SADS-CoV plays a vital role in viral assembly and replication, but its structure and functions remain poorly understood. This study utilized biochemistry, X-ray crystallography, and immunization techniques to investigate the N protein's structure and function in SADS-CoV. Our findings revealed distinct domains within the N protein, including an RNA-binding domain, two disordered domains, and a dimerization domain. Through biochemical assays, we confirmed that the N-terminal domain functions as an RNA-binding domain, and the C-terminal domain is involved in dimerization, with the crystal structure analysis providing visual evidence of dimer formation. Immunization experiments demonstrated that the disordered domain 2 elicited a significant antibody response. These identified domains and their interactions are crucial for viral assembly. This comprehensive understanding of the N protein in SADS-CoV enhances our knowledge of its assembly and replication mechanisms, enabling the development of targeted interventions and therapeutic strategies. SADS-CoV is a porcine coronavirus that originated from a bat HKU2-related coronavirus. It causes devastating swine diseases and poses a high risk of spillover to humans. The coronavirus N protein, as the most abundant viral protein in infected cells, likely plays a key role in viral assembly and replication. However, the structure and function of this protein remain unclear. Therefore, this study employed a combination of biochemistry and X-ray crystallography to uncover distinct structural domains in the N protein, including RNA-binding domains, two disordered domains, and dimerization domains. Additionally, we made the novel discovery that the disordered domain elicited a significant antibody response. These findings provide new insights into the structure and functions of the SADS-CoV N protein, which have important implications for future studies on SADS-CoV diagnosis, as well as the development of vaccines and anti-viral drugs.
Organizational Affiliation:
Guangdong Provincial Key Laboratory of New Drug Screening & NMPA Key Laboratory for Research and Evaluation of Drug Metabolism & Guangdong-Hong Kong-Macao Joint Laboratory for New Drug Screening, School of Pharmaceutical Sciences, Southern Medical University, Guangzhou, China.