Molecular Basis of VCPIP1 and P97/VCP Interaction Reveals Its Functions in Post-Mitotic Golgi Reassembly.
Liao, T., Li, R., Lu, P., Liu, Y., Yang, R., Guo, H., Wu, Z., Wang, R., Yuan, L., Hu, Z., Gao, H., Li, F.(2024) Adv Sci (Weinh) 11: e2403417-e2403417
- PubMed: 39234822
- DOI: https://doi.org/10.1002/advs.202403417
- Primary Citation of Related Structures:
8YKA - PubMed Abstract:
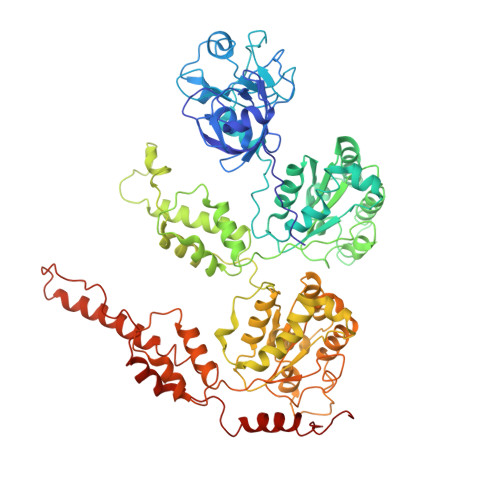
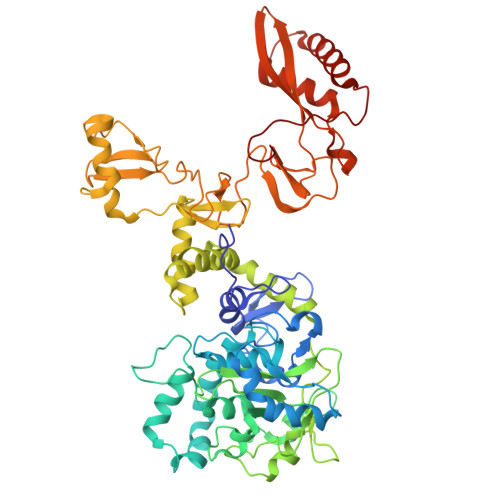
The VCPIP1-P97/VCP (Valosin-Containing Protein) complex is required for post-mitotic Golgi cisternae reassembly and maintenance in interphase. However, the organization and mechanism of this complex in regulating Golgi membrane fusion is still elusive. Here, the cryo-electron microscopy (cryo-EM) structures of the human VCPIP1-P97/VCP complex are presented. These studies reveal that three independent VCPIP1 molecules sit over the C-terminal substrate exit tunnel formed by P97/VCP homo-hexamer, resulting in an unusual C3 to C6 symmetric barrel architecture. The UFD1 (unknown function domain 1) from VCPIP1, but not the N-terminal OTU domain and the C-terminal UBL domain, docks to the two adjacent D2 domains of P97/VCP, allosterically causing the cofactors binding domain-NTDs (N-terminal domains) of P97/VCP in a "UP" and D1 domain in an ATPase competent conformation. Conversely, VCPIP1 bound P97/VCP hexamer favors the binding of P47, and thus the intact SNARE complex, promoting Golgi membrane fusion. These studies not only reveal the unexpected organization of humanVCPIP1-P97/VCP complex, but also provide new insights into the mechanism of VCPIP1-P97/VCP mediated Golgi apparatus reassembly, which is a fundamental cellular event for protein and lipid processing.
- MOE Key Laboratory of Rare Pediatric Diseases, Center for Medical Genetics, School of Life Sciences, Central South University, Changsha, Hunan, 410013, China.
Organizational Affiliation: