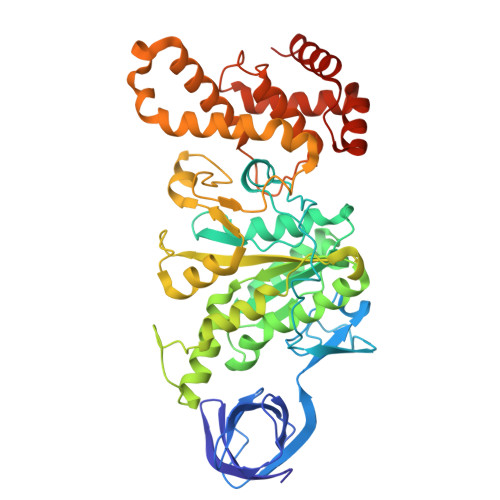
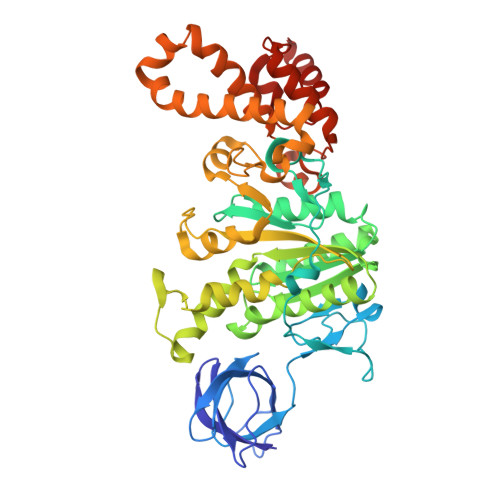
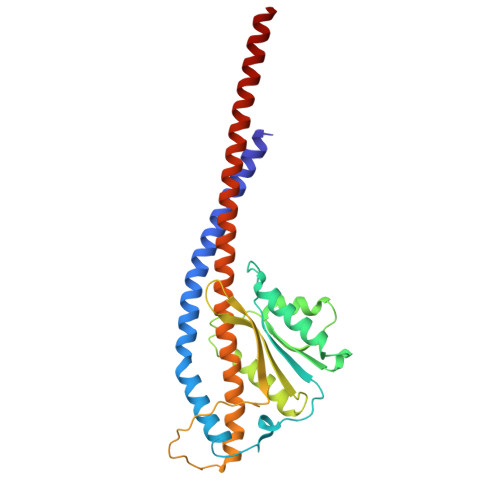
ADP-inhibited structure of non-catalytic site-depleted F o F 1 -ATPase from thermophilic Bacillus sp. PS-3.
Kobayashi, R., Nakano, A., Mitsuoka, K., Yokoyama, K.(2025) Biochim Biophys Acta Bioenerg 1866: 149536-149536
- PubMed: 39788275
- DOI: https://doi.org/10.1016/j.bbabio.2025.149536
- Primary Citation of Related Structures:
8YGV, 8YH8 - PubMed Abstract:
The F 1 domain of F o F 1 -ATP synthases/ATPases (F o F 1 ) possesses three catalytic sites on the three αβ interfaces, termed α E β E , α D β D , and α T β T , located mainly on the β subunits. The enzyme also has three non-catalytic ATP-binding sites on the three αβ interfaces, located mainly on the α subunits. When ATP does not bind to the non-catalytic site, F o F 1 becomes significantly prone to ADP inhibition, ultimately resulting in the loss of ATPase activity. However, the underlying mechanism of ADP inhibition remains unclear. Here, we report the cryo-EM structure of the non-catalytic site-depleted (ΔNC) F o F 1 from thermophilic Bacillus sp. PS-3, which completely lacks the ability to bind ATP (and ADP) upon transitioning to the ADP-inhibited form. The structure closely resembled the 81° rotated structure of the wild-type F o F 1 , except for minor movements in the C-terminal region of the α subunit. In this structure, unlike the wild-type enzyme, the catalytic site at α D β D , responsible for ATP hydrolysis, was occupied by ADP-Mg, with the absence of Pi. Furthermore, the catalytic site at α E β E , where ATP enters the F 1 domain during steady-state catalysis, is occupied by ADP, seemingly impeding further ATP binding to the enzyme. The structure suggests that the ADP-inhibited form of the F 1 domain is more likely due to differences in the nucleotide-binding states at the catalytic sites rather than structural differences.
- Department of Molecular Biosciences, Kyoto Sangyo University, Kamigamo-Motoyama, Kita-ku, Kyoto 603-8555, Japan.
Organizational Affiliation: