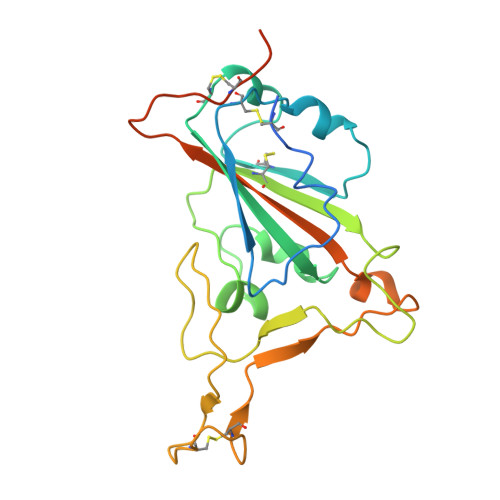
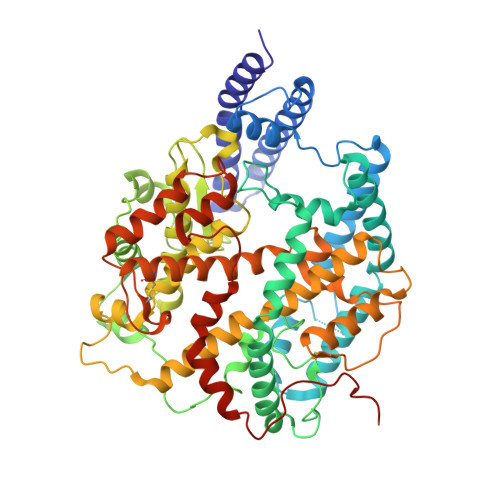
Receptor binding and structural basis of raccoon dog ACE2 binding to SARS-CoV-2 prototype and its variants.
Luo, C., Li, L., Gu, Y., Zhang, H., Xu, Z., Sun, J., Shi, K., Ma, S., Tian, W.X., Liu, K., Gao, G.F.(2024) PLoS Pathog 20: e1012713-e1012713
- PubMed: 39637248
- DOI: https://doi.org/10.1371/journal.ppat.1012713
- Primary Citation of Related Structures:
8YF2, 8YFT - PubMed Abstract:
Raccoon dog was proposed as a potential host of SARS-CoV-2, but no evidence support such a notion. In our study, we investigated the binding affinities of raccoon dog ACE2 (rdACE2) to the spike (S) protein receptor binding domain (RBD) of SARS-CoV-2 prototype (PT) and its variants. It revealed that the binding affinities of RBD from SARS-CoV-2 variants were generally lower than that of the PT RBD. Through structural and functional analyses, we found amino acids H34 and M82 play pivotal roles in maintaining the binding affinity of ACE2 to different SARS-CoV-2 sub-variants. These results suggest that raccoon dogs exhibit lower susceptibility to SARS-CoV-2 compared to those animal species with a high prevalence of SARS-CoV-2 transmission.
- College of Veterinary Medicine, Shanxi Agricultural University, Jinzhong, China.
Organizational Affiliation: