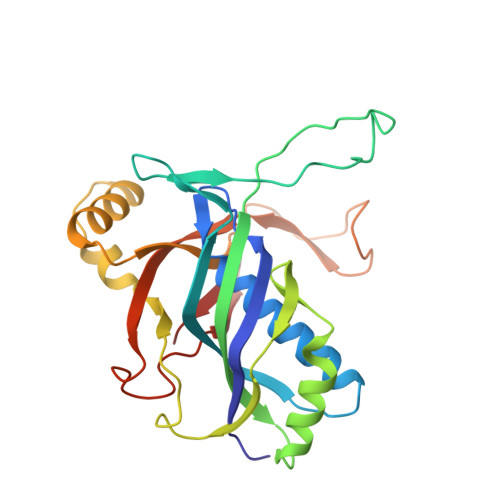
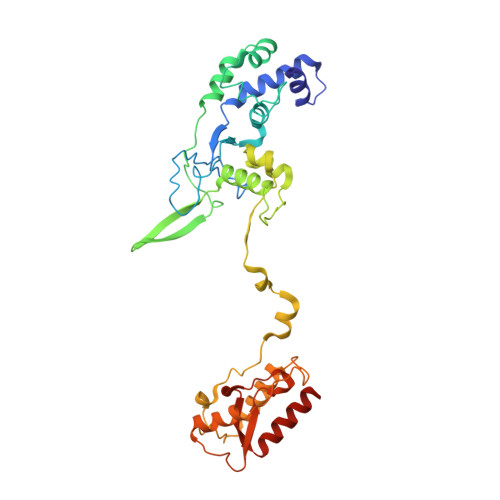
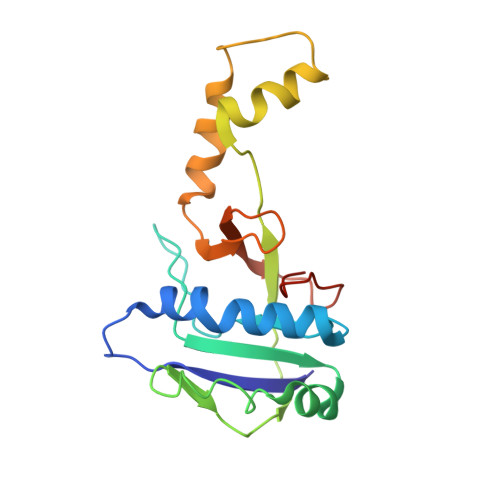
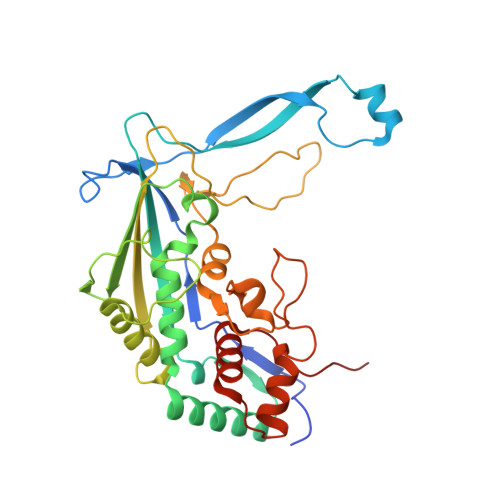
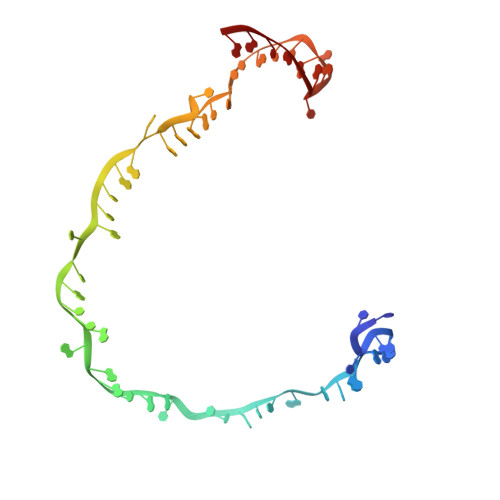
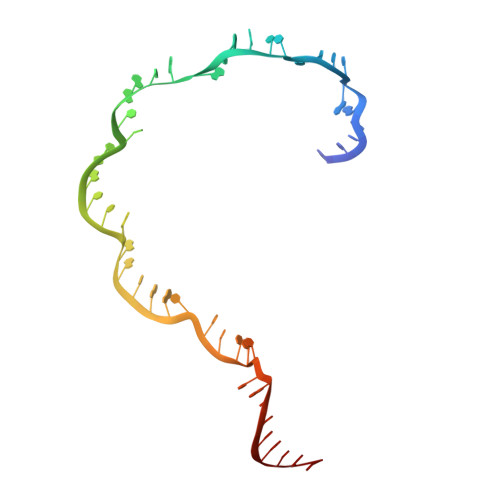
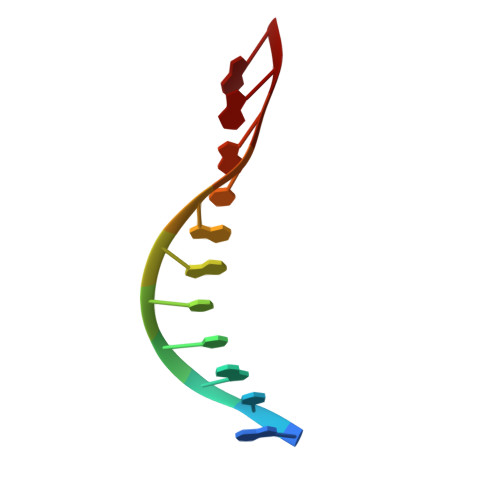
Mechanisms for HNH-mediated target DNA cleavage in type I CRISPR-Cas systems.
Zhang, C., Chen, F., Wang, F., Xu, H., Xue, J., Li, Z.(2024) Mol Cell 84: 3141
- PubMed: 39047725
- DOI: https://doi.org/10.1016/j.molcel.2024.06.033
- Primary Citation of Related Structures:
8YB6, 8YDB, 8YEO, 8YH9, 8YHA - PubMed Abstract:
The metagenome-derived type I-E and type I-F variant CRISPR-associated complex for antiviral defense (Cascade) complexes, fused with HNH domains, precisely cleave target DNA, representing recently identified genome editing tools. However, the underlying working mechanisms remain unknown. Here, structures of type I-F HNH and I-E HNH Cascade complexes at different states are reported. In type I-F HNH Cascade, Cas8f HNH loosely attaches to Cascade head and is adjacent to the 5' end of the target single-stranded DNA (ssDNA). Formation of the full R-loop drives the Cascade head to move outward, allowing Cas8f HNH to detach and rotate ∼150° to accommodate target ssDNA for cleavage. In type I-E HNH Cascade, Cas5e HNH domain is adjacent to the 5' end of the target ssDNA. Full crRNA-target pairing drives the lift of the Cascade head, widening the substrate channel for target ssDNA entrance. Altogether, these analyses into both complexes revealed that crRNA-guided positioning of target DNA and target DNA-induced HNH unlocking are two key factors for their site-specific cleavage of target DNA.
- State Key Laboratory of Biocatalysis and Enzyme Engineering, School of Life Sciences, Hubei University, Wuhan 430062, Hubei, China.
Organizational Affiliation: