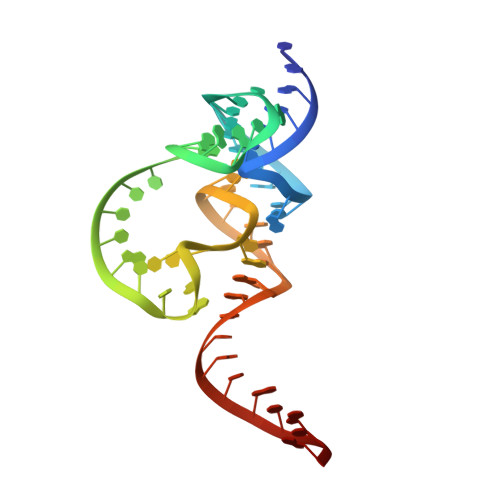
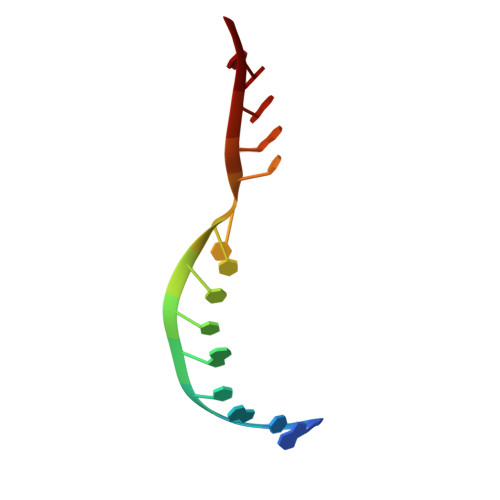
The structure and catalytic mechanism of a pseudoknot-containing hammerhead ribozyme.
Zhan, X., Wilson, T.J., Li, Z., Zhang, J., Yang, Y., Lilley, D.M.J., Liu, Y.(2024) Nat Commun 15: 6628-6628
- PubMed: 39103372
- DOI: https://doi.org/10.1038/s41467-024-50892-y
- Primary Citation of Related Structures:
8YDC - PubMed Abstract:
We have determined the crystal structure of a pseudoknot (PK)-containing hammerhead ribozyme that closely resembles the pistol ribozyme, with essentially identical secondary structure and connectivity. The activity is more sensitive to deletion of the G8 2'OH than to the absence of magnesium ions, indicating that the catalytic mechanism is the same as the extended hammerhead, and distinct from the pistol ribozyme. Here we show that nucleophilic attack is almost perfectly in-line, and the G8 2'OH is well positioned to act as general acid, being directed towards the O5' leaving group, and 2.9 Å away from it. Despite the similarity in overall structure to the pistol ribozyme, the local structure close to the cleavage site differs, and the PK hammerhead retains its unique mechanistic identity and demonstrates enhanced activity over other hammerhead ribozymes under standard conditions.
- State Key Laboratory of Medicinal Chemical Biology, Frontiers Science Center for Cell Responses, Nankai University, Tianjin, China.
Organizational Affiliation: