Deciphering DED assembly mechanisms in FADD-procaspase-8-cFLIP complexes regulating apoptosis.
Yang, C.Y., Lien, C.I., Tseng, Y.C., Tu, Y.F., Kulczyk, A.W., Lu, Y.C., Wang, Y.T., Su, T.W., Hsu, L.C., Lo, Y.C., Lin, S.C.(2024) Nat Commun 15: 3791-3791
- PubMed: 38710704
- DOI: https://doi.org/10.1038/s41467-024-47990-2
- Primary Citation of Related Structures:
8YBX, 8YD7, 8YD8 - PubMed Abstract:
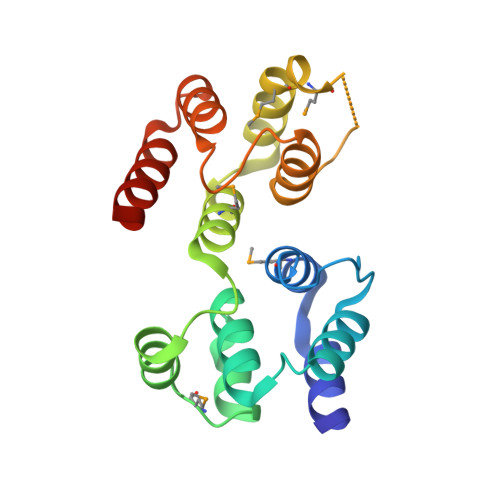
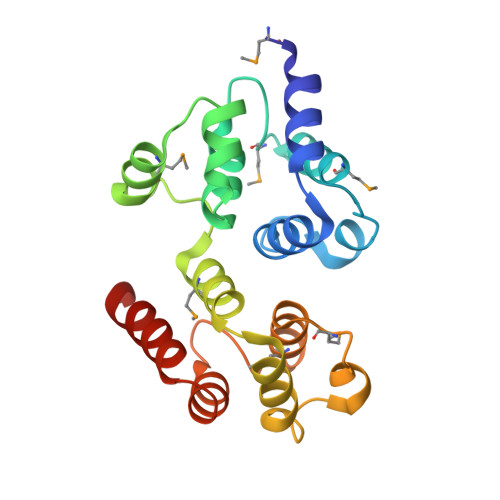
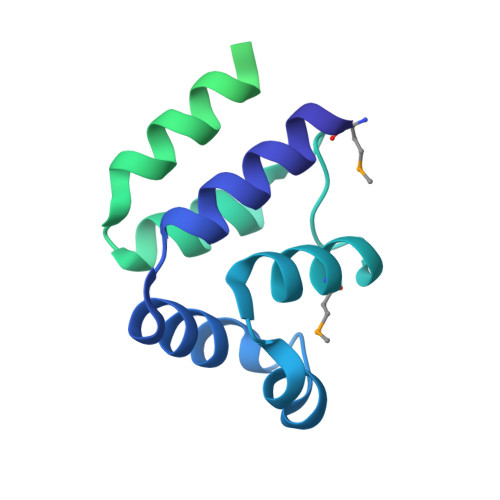
Fas-associated protein with death domain (FADD), procaspase-8, and cellular FLICE-inhibitory proteins (cFLIP) assemble through death-effector domains (DEDs), directing death receptor signaling towards cell survival or apoptosis. Understanding their three-dimensional regulatory mechanism has been limited by the absence of atomic coordinates for their ternary DED complex. By employing X-ray crystallography and cryogenic electron microscopy (cryo-EM), we present the atomic coordinates of human FADD-procaspase-8-cFLIP complexes, revealing structural insights into these critical interactions. These structures illustrate how FADD and cFLIP orchestrate the assembly of caspase-8-containing complexes and offer mechanistic explanations for their role in promoting or inhibiting apoptotic and necroptotic signaling. A helical procaspase-8-cFLIP hetero-double layer in the complex appears to promote limited caspase-8 activation for cell survival. Our structure-guided mutagenesis supports the role of the triple-FADD complex in caspase-8 activation and in regulating receptor-interacting protein kinase 1 (RIPK1). These results propose a unified mechanism for DED assembly and procaspase-8 activation in the regulation of apoptotic and necroptotic signaling across various cellular pathways involved in development, innate immunity, and disease.
- Genomics Research Center, Academia Sinica, Taipei, 11529, Taiwan.
Organizational Affiliation: