Activation mechanism of caseinolytic chaperone-protease system in Mycobacterium tuberculosis by the anti-cancer drug bortezomib
Zhou, B., Zhao, H., Gao, Y., Chen, X., Zhang, T., He, J., Xiong, X.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ATP-dependent Clp protease ATP-binding subunit ClpC1 | 657 | Mycobacterium tuberculosis H37Rv | Mutation(s): 3 Gene Names: clpC1 | 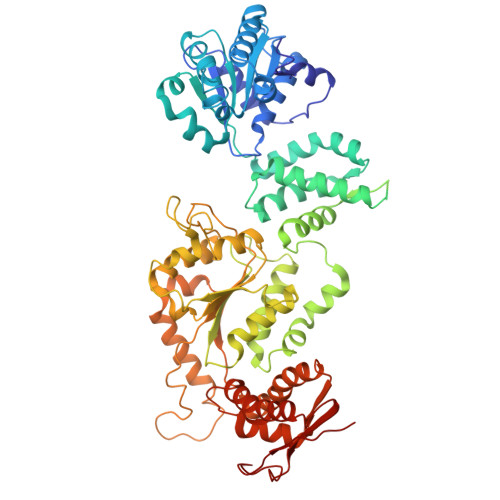 | |
UniProt | |||||
Find proteins for P9WPC9 (Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv)) Explore P9WPC9 Go to UniProtKB: P9WPC9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P9WPC9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ATP-dependent Clp protease proteolytic subunit 2 | 195 | Mycobacterium tuberculosis H37Rv | Mutation(s): 0 Gene Names: clpP2 EC: 3.4.21.92 | 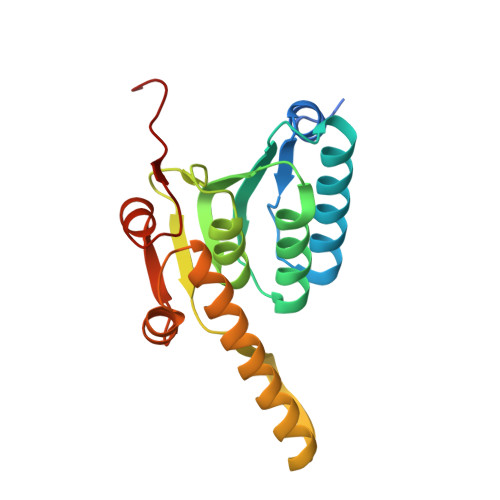 | |
UniProt | |||||
Find proteins for P9WPC3 (Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv)) Explore P9WPC3 Go to UniProtKB: P9WPC3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P9WPC3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ATP-dependent Clp protease proteolytic subunit 1 | 178 | Mycobacterium tuberculosis H37Rv | Mutation(s): 0 Gene Names: clpP1, clpP EC: 3.4.21.92 | 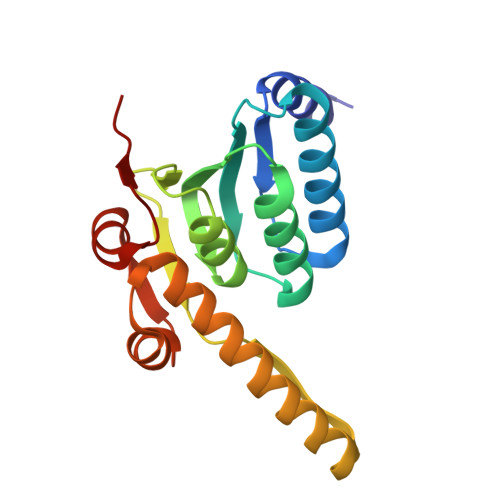 | |
UniProt | |||||
Find proteins for P9WPC5 (Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv)) Explore P9WPC5 Go to UniProtKB: P9WPC5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P9WPC5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Beta-casein | 23 | Bos grunniens | Mutation(s): 0 | 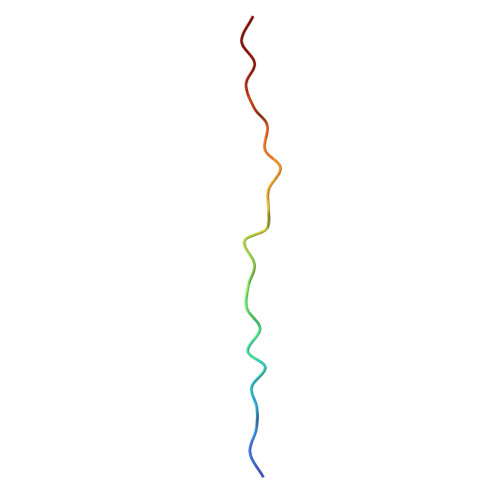 | |
UniProt | |||||
Find proteins for P02666 (Bos taurus) Explore P02666 Go to UniProtKB: P02666 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P02666 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ATP (Subject of Investigation/LOI) Query on ATP | BA [auth B] CA [auth B] FA [auth C] GA [auth C] LA [auth E] | ADENOSINE-5'-TRIPHOSPHATE C10 H16 N5 O13 P3 ZKHQWZAMYRWXGA-KQYNXXCUSA-N |  | ||
| ADP (Subject of Investigation/LOI) Query on ADP | IA [auth D], KA [auth E], MA [auth F], V [auth A] | ADENOSINE-5'-DIPHOSPHATE C10 H15 N5 O10 P2 XTWYTFMLZFPYCI-KQYNXXCUSA-N |  | ||
| BO2 (Subject of Investigation/LOI) Query on BO2 | AB [auth T] NA [auth G] OA [auth H] PA [auth I] QA [auth J] | N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE C19 H25 B N4 O4 GXJABQQUPOEUTA-RDJZCZTQSA-N |  | ||
| MG (Subject of Investigation/LOI) Query on MG | AA [auth B] DA [auth B] EA [auth C] HA [auth C] JA [auth D] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Funding Organization | Location | Grant Number |
|---|---|---|
| Other government | 2021YFA1300903 | |
| Other government | 2021YFA1300903 | |
| National Natural Science Foundation of China (NSFC) | China | 32300152 |
| National Natural Science Foundation of China (NSFC) | China | 81973372 |
| National Natural Science Foundation of China (NSFC) | China | 32170189 |
| National Natural Science Foundation of China (NSFC) | China | 32241021 |
| National Natural Science Foundation of China (NSFC) | China | 82341085 |
| Other government | 2022A1515110505 | |
| Other government | 2022M723164 |