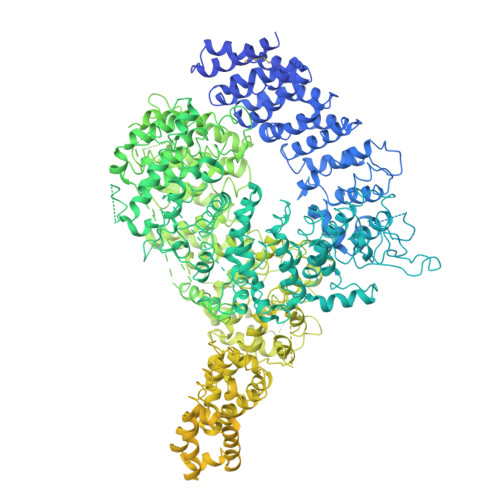
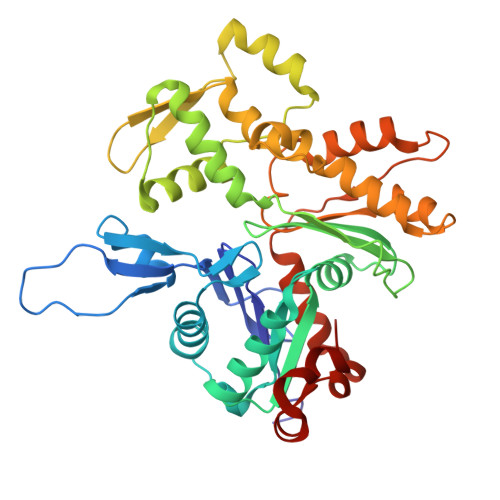
Structure of the Huntingtin F-actin complex reveals its role in cytoskeleton organization.
Carpentier, R., Kim, J., Capizzi, M., Kim, H., Fassler, F., Hansen, J.M., Kim, M.J., Denarier, E., Blot, B., Degennaro, M., Labou, S., Arnal, I., Marcaida, M.J., Peraro, M.D., Kim, D., Schur, F.K.M., Song, J.J., Humbert, S.(2025) Sci Adv 11: eadw4124-eadw4124
- PubMed: 40971423
- DOI: https://doi.org/10.1126/sciadv.adw4124
- Primary Citation of Related Structures:
8YAE, 8YAO - PubMed Abstract:
The Huntingtin protein (HTT), named for its role in Huntington's disease, has been best understood as a scaffolding protein that promotes vesicle transport by molecular motors along microtubules. Here, we show that HTT also interacts with the actin cytoskeleton, and its loss of function disturbs the morphology and function of the axonal growth cone. We demonstrate that HTT organizes F-actin into bundles. Cryo-electron tomography (cryo-ET) and subtomogram averaging (STA) structural analyses reveal that HTT's N-terminal HEAT and Bridge domains wrap around F-actin, while the C-terminal HEAT domain is displaced; furthermore, HTT dimerizes via the N-HEAT domain to bridge parallel actin filaments separated by ~20 nanometers. Our study provides the structural basis for understanding how HTT interacts with and organizes the actin cytoskeleton.
- Univ. Grenoble Alpes, Inserm, U1216, CEA, Grenoble Institute Neurosciences, 38000 Grenoble, France.
Organizational Affiliation: