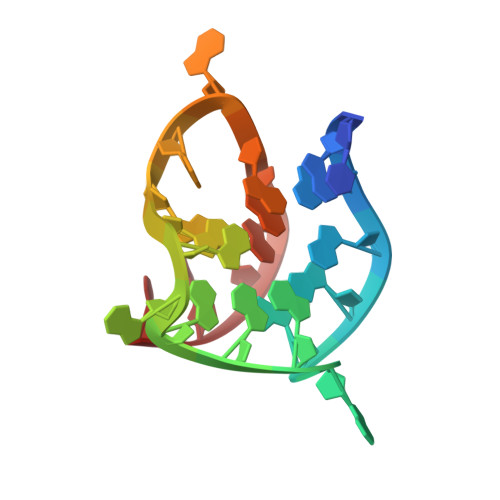
Overcoming Cancer Persister Cells by Stabilizing the ATF4 Promoter G-quadruplex.
Xiao, C., Li, Y., Liu, Y., Dong, R., He, X., Lin, Q., Zang, X., Wang, K., Xia, Y., Kong, L.(2024) Adv Sci (Weinh) 11: e2401748-e2401748
- PubMed: 38994891
- DOI: https://doi.org/10.1002/advs.202401748
- Primary Citation of Related Structures:
8Y2R - PubMed Abstract:
Persister cells (PS) selected for anticancer therapy have been recognized as a significant contributor to the development of treatment-resistant malignancies. It is found that imposing glutamine restriction induces the generation of PS, which paradoxically bestows heightened resistance to glutamine restriction treatment by activating the integrated stress response and initiating the general control nonderepressible 2-activating transcription factor 4-alanine, serine, cysteine-preferring transporter 2 (GCN2-ATF4-ASCT2) axis. Central to this phenomenon is the stress-induced ATF4 translational reprogramming. Unfortunately, directly targeting ATF4 protein has proven to be a formidable challenge because of its flat surface. Nonetheless, a G-quadruplex structure located within the promoter region of ATF4 (ATF4-G4) is uncovered and resolved, which functions as a transcriptional regulator and can be targeted by small molecules. The investigation identifies the natural compound coptisine (COP) as a potent binder that interacts with and stabilizes ATF4-G4. For the first time, the high-resolution structure of the COP-ATF4-G4 complex is determined. The formation of this stable complex disrupts the interaction between transcription factor AP-2 alpha (TFAP2A) and ATF4-G4, resulting in a substantial reduction in intracellular ATF4 levels and the eventual death of cancer cells. These seminal findings underscore the potential of targeting the ATF4-G4 structure to yield significant therapeutic advantages within the realm of persister cancer cells induced by glutamine-restricted therapy.
- State Key Laboratory of Natural Medicines and Jiangsu Key Laboratory of Bioactive Natural Product Research, School of Traditional Chinese Pharmacy, China Pharmaceutical University, Nanjing, 210009, China.
Organizational Affiliation: