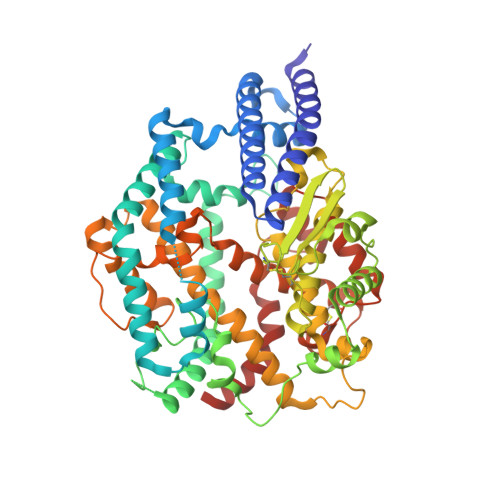
Mosaic RBD nanoparticle elicits immunodominant antibody responses across sarbecoviruses.
Liu, C., Xu, S., Zheng, Y., Xie, Y., Xu, K., Chai, Y., Luo, T., Dai, L., Gao, G.F.(2024) Cell Rep 43: 114235-114235
- PubMed: 38748880
- DOI: https://doi.org/10.1016/j.celrep.2024.114235
- Primary Citation of Related Structures:
8XXW - PubMed Abstract:
Nanoparticle vaccines displaying mosaic receptor-binding domains (RBDs) or spike (S) from severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) or other sarbecoviruses are used in preparedness against potential zoonotic outbreaks. Here, we describe a self-assembling nanoparticle using lumazine synthase (LuS) as the scaffold to display RBDs from different sarbecoviruses. Mosaic nanoparticles induce sarbecovirus cross-neutralizing antibodies comparable to a nanoparticle cocktail. We find mosaic nanoparticles elicit a B cell receptor repertoire using an immunodominant germline gene pair of IGHV14-3:IGKV14-111. Most of the tested IGHV14-3:IGKV14-111 monoclonal antibodies (mAbs) are broadly cross-reactive to clade 1a, 1b, and 3 sarbecoviruses. Using mAb competition and cryo-electron microscopy, we determine that a representative IGHV14-3:IGKV14-111 mAb, M2-7, binds to a conserved epitope on the RBD, largely overlapping with the pan-sarbecovirus mAb S2H97. This suggests mosaic nanoparticles expand B cell recognition of the common epitopes shared by different clades of sarbecoviruses. These results provide immunological insights into the cross-reactive responses elicited by mosaic nanoparticles against sarbecoviruses.
- College of Animal Science and Veterinary Medicine, Guangxi University, Nanning 530004, Guangxi, China; CAS Key Laboratory of Pathogen Microbiology and Immunology, Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, China.
Organizational Affiliation: