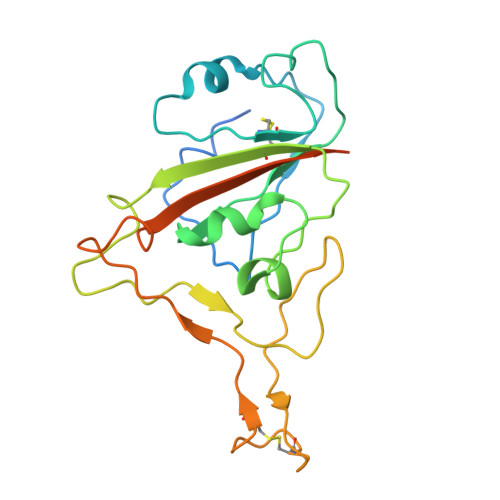
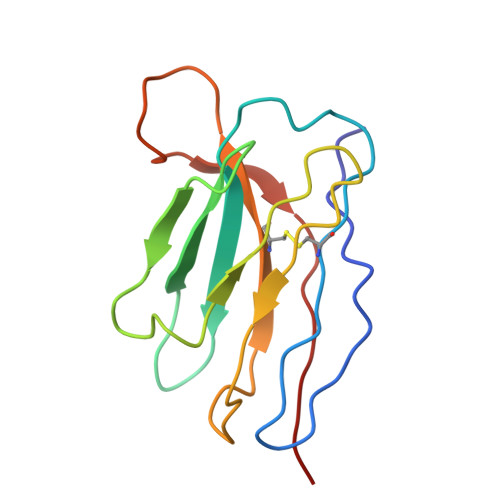
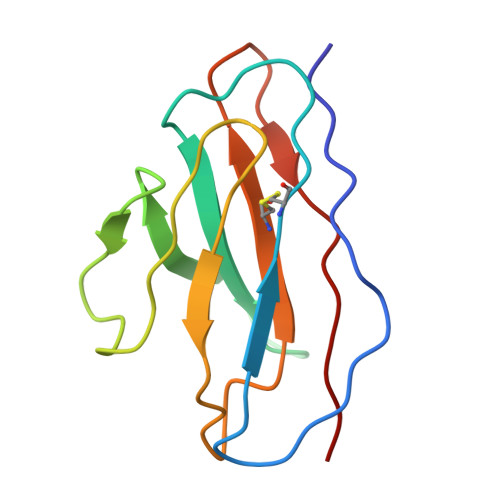
Rapid clonal expansion and somatic hypermutation contribute to the fate of SARS-CoV-2 broadly neutralizing antibodies.
Wang, M., Liu, C., Fan, Q., Sun, Y., Tang, S., Guo, H., Zhou, B., Wang, H., Ge, X., Zhang, Z., Ju, B.(2025) J Immunol 214: 278-289
- PubMed: 40073246
- DOI: https://doi.org/10.1093/jimmun/vkae056
- Primary Citation of Related Structures:
8XRQ - PubMed Abstract:
Several vaccines and immunization strategies, including inactivated vaccines, have proven effective in eliciting antibodies against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), providing an opportunity to characterize the antibody response. In this study, we investigated the monoclonal antibody responses elicited by wild-type SARS-CoV-2 inactivated vaccination compared to those elicited by natural infection and mRNA vaccination. The analysis showed that antibodies encoded by biased germline genes were shared between SARS-CoV-2 vaccinated and naturally infected individuals. Among the 35 shared clonotypes identified, besides the well-known IGHV3-53 and IGHV1-58, we identified a class of IGHV4-59 antibodies characterized by rapid response and neutralizing activity, elicited by 3 doses of inactivated vaccine. Members of this lineage exhibited similar sensitivity against wild-type SARS-CoV-2, whereas different neutralizing activities against SARS-CoV-2 variants, especially against various Omicron subvariants, BA.1, BA.2, BA.2.12.1, BA.4/5, and BA.2.75. Structural analysis of BA.1 spike complexed with VacBB-639 revealed that the IGHV4-59-lineage antibodies belonged to the Class 2/3 group. Using sequence alignment, site-mutation assays, and functional verification, we identified two substitutions, N60K in HFR3 and S56G in HCDR2, contributing to opposite neutralization changes of IGHV4-59-lineage antibodies against these Omicron subvariants. These results demonstrate the importance of somatic hypermutation in the evolution of prototypical antigen-elicited antibodies in terms of their neutralization breadth and potency against SARS-CoV-2 Omicron variants.
- Institute for Hepatology, National Clinical Research Center for Infectious Disease, Shenzhen Third People's Hospital, The Second Affiliated Hospital, School of Medicine, Southern University of Science and Technology, Shenzhen, Guangdong Province, China.
Organizational Affiliation: