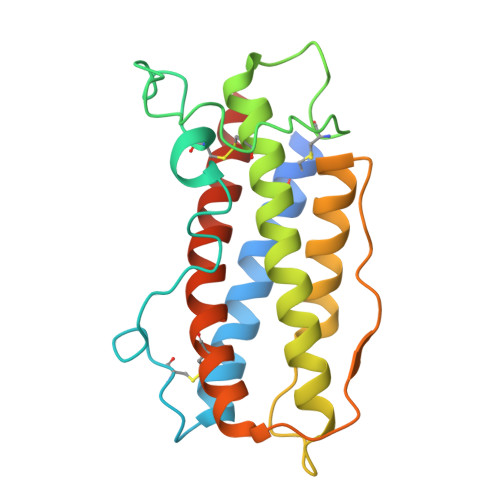
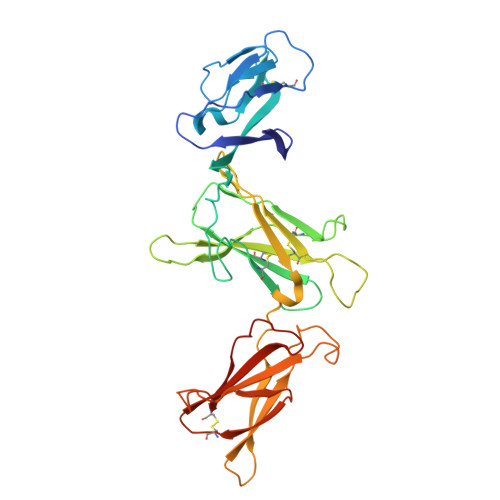
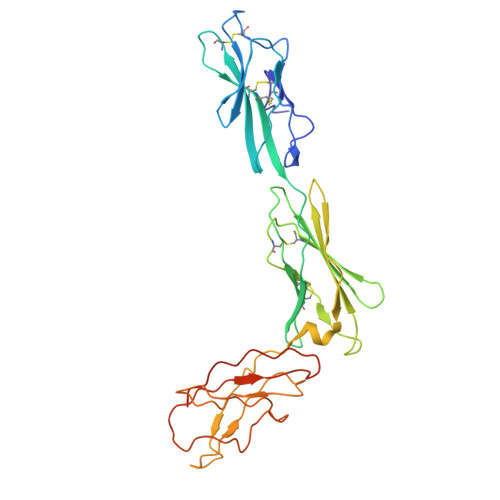
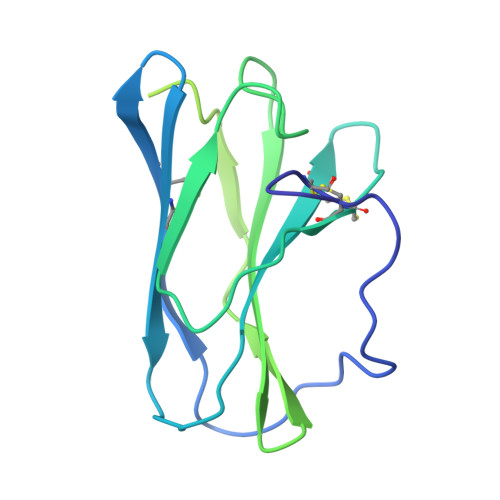
Structure and assembly of the human IL-12 signaling complex.
Chen, H., Ge, X., Li, C., Zeng, J., Wang, X.(2024) Structure 32: 1640
- PubMed: 39111304
- DOI: https://doi.org/10.1016/j.str.2024.07.010
- Primary Citation of Related Structures:
8XRP, 8YI7 - PubMed Abstract:
Interleukin (IL)-12 is a heterodimeric pro-inflammatory cytokine. Our cryoelectron microscopy structure determination of human IL-12 in complex with IL-12Rβ1 and IL-12Rβ2 at a resolution of 3.75 Å reveals that IL-12Rβ2 primarily interacts with the IL-12p35 subunit via its N-terminal Ig-like domain, while IL-12Rβ1 binds to the p40 subunit with its N-terminal fibronectin III domain. This binding mode of IL-12 with its receptors is similar to that of IL-23 but shows notable differences with other cytokines. Through structural information and biochemical assays, we identified Y62, Y189, and K192 as key residues in IL-12p35, which bind to IL-12Rβ2 with high affinity and mediate IL-12 signal transduction. Furthermore, structural comparisons reveal two distinctive conformational states and structural plasticity of the heterodimeric interface in IL-12. As a result, our study advances our understanding of IL-12 signal initiation and opens up new opportunities for the engineering and therapeutic targeting of IL-12.
- The Ministry of Education Key Laboratory of Protein Science, Beijing Frontier Research Center for Biological Structure, School of Life Sciences, Tsinghua University, Beijing 100084, China.
Organizational Affiliation: