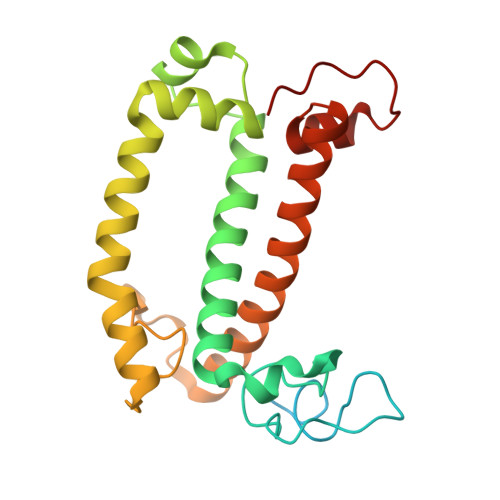
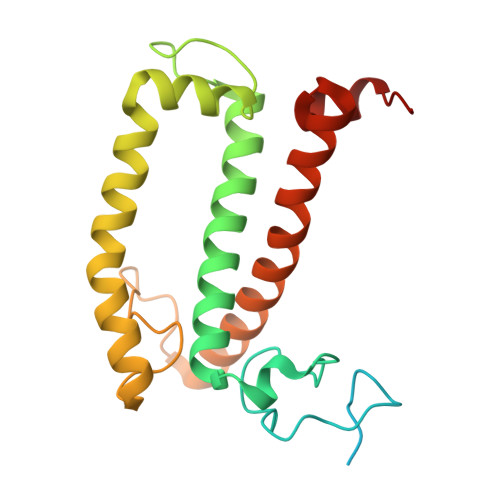
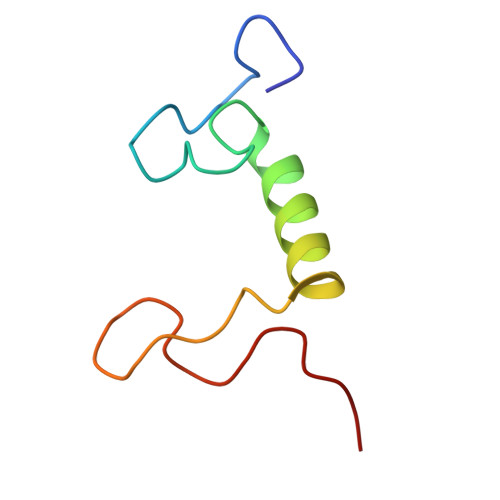
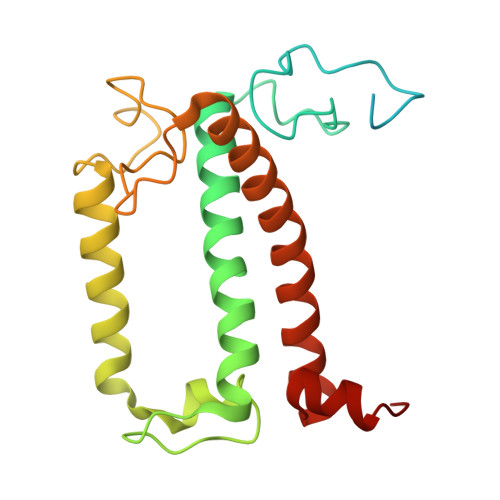
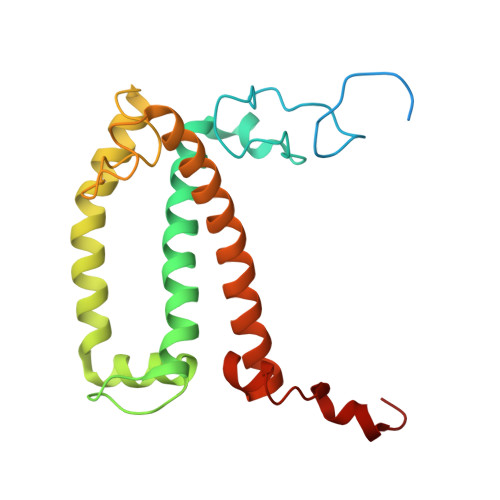
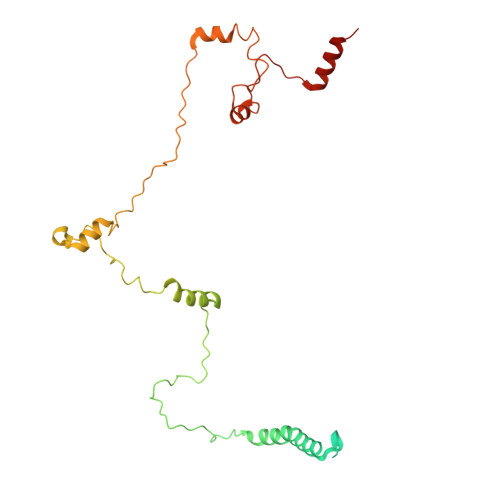
Structure and distinct supramolecular organization of a PSII-ACPII dimer from a cryptophyte alga Chroomonas placoidea.
Mao, Z., Li, X., Li, Z., Shen, L., Li, X., Yang, Y., Wang, W., Kuang, T., Shen, J.R., Han, G.(2024) Nat Commun 15: 4535-4535
- PubMed: 38806516
- DOI: https://doi.org/10.1038/s41467-024-48878-x
- Primary Citation of Related Structures:
8WB4, 8XKL - PubMed Abstract:
Cryptophyte algae are an evolutionarily distinct and ecologically important group of photosynthetic unicellular eukaryotes. Photosystem II (PSII) of cryptophyte algae associates with alloxanthin chlorophyll a/c-binding proteins (ACPs) to act as the peripheral light-harvesting system, whose supramolecular organization is unknown. Here, we purify the PSII-ACPII supercomplex from a cryptophyte alga Chroomonas placoidea (C. placoidea), and analyze its structure at a resolution of 2.47 Å using cryo-electron microscopy. This structure reveals a dimeric organization of PSII-ACPII containing two PSII core monomers flanked by six symmetrically arranged ACPII subunits. The PSII core is conserved whereas the organization of ACPII subunits exhibits a distinct pattern, different from those observed so far in PSII of other algae and higher plants. Furthermore, we find a Chl a-binding antenna subunit, CCPII-S, which mediates interaction of ACPII with the PSII core. These results provide a structural basis for the assembly of antennas within the supercomplex and possible excitation energy transfer pathways in cryptophyte algal PSII, shedding light on the diversity of supramolecular organization of photosynthetic machinery.
- Photosynthesis Research Center, Key Laboratory of Photobiology, Institute of Botany, Chinese Academy of Sciences, 100093, Beijing, China.
Organizational Affiliation: