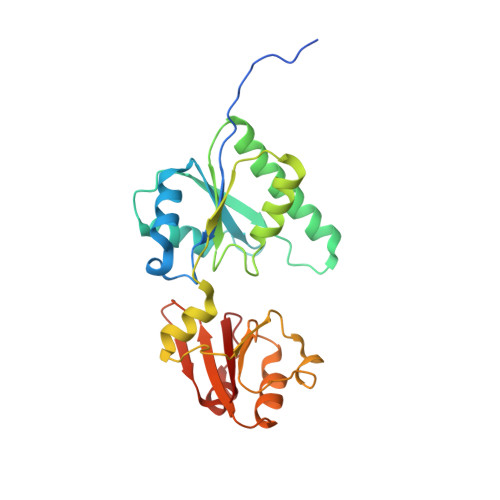
Crystal structure of methyltransferase CbiL from Akkermansia muciniphila.
Guo, S., Jiang, M., Wang, W., Chen, X., Wei, Q., Wang, M.(2024) Biochem Biophys Res Commun 722: 150165-150165
- PubMed: 38805786
- DOI: https://doi.org/10.1016/j.bbrc.2024.150165
- Primary Citation of Related Structures:
8XJ3 - PubMed Abstract:
Akkermansia muciniphila is a mucin-degrading probiotic that colonizes the gastrointestinal tract. Genomic analysis identified a set of genes involved in the biosynthesis of corrin ring, including the cobalt factor II methyltransferase CbiL, in some phylogroups of A. muciniphila, implying a potential capacity for de novo synthesis of cobalamin. In this work, we determined the crystal structure of CbiL from A. muciniphila at 2.3 Å resolution. AmCbiL exists as a dimer both in solution and in crystal, and each protomer consists of two α/β domains, the N-terminal domain and the C-terminal domain, consistent with the folding of typical class III MTases. The two domains create an open trough, potentially available to bind the substrates SAM and cobalt factor II. Sequence and structural comparisons with other CbiLs, assisted by computer modeling, suggest that AmCbiL should have cobalt factor II C-20 methyltransferase activity. Our results support that certain strains of A. muciniphila may be capable of synthesizing cobalamin de novo.
Organizational Affiliation:
Institutes of Physical Science and Information Technology, Anhui University, Hefei 230601, Anhui, China; School of Life Sciences, Anhui University, Hefei 230601, Anhui, China.