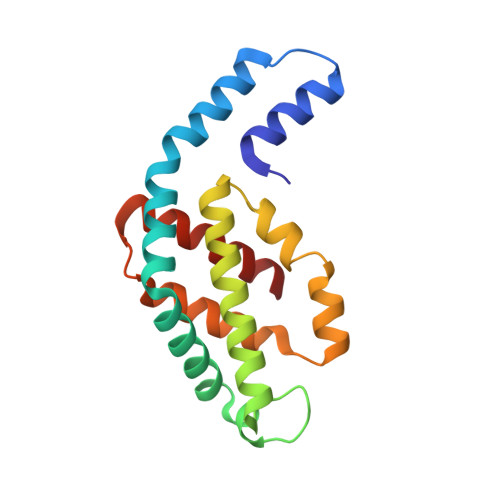
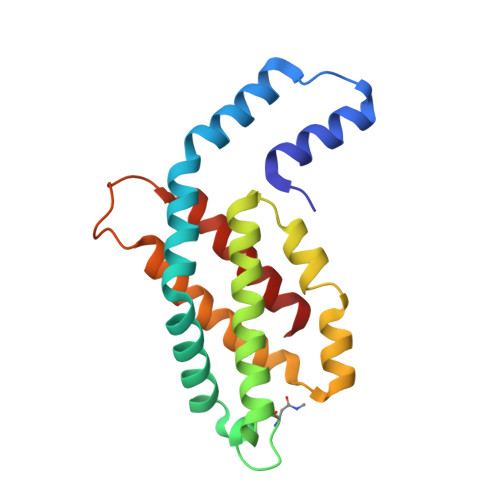
Disassembly and reassembly of the non-conventional thermophilic C-phycocyanin.
Nguyen, H.K., Minato, T., Teramoto, T., Ogo, S., Kakuta, Y., Yoon, K.S.(2024) J Biosci Bioeng 137: 179-186
- PubMed: 38238241
- DOI: https://doi.org/10.1016/j.jbiosc.2023.12.015
- Primary Citation of Related Structures:
8XF0, 8XF1, 8XF2, 8XF3 - PubMed Abstract:
C-phycocyanin (CPC), which contains open-chain tetrapyrroles, is a major light-harvesting red-fluorescent protein with an important role in aquatic photosynthesis. Recently, we reported a non-conventional CPC from Thermoleptolyngbya sp. O-77 (CPC O77 ) that contains two different structures, i.e., a hexameric structure and a non-conventional octameric structure. However, the assembly and disassembly mechanisms of the non-conventional octameric form of CPC remain unclear. To understand this assembly mechanism, we performed an in vitro experiment to study the disassembly and reassembly behaviors of CPC using isolated CPC subunits. The dissociation of the CPC O77 subunit was performed using a Phenyl-Sepharose column in 20 mM potassium phosphate buffer (pH 6.0) containing 7.0 M urea. For the first time, crystals of isolated CPC subunits were obtained and analyzed after separation. After the removal of urea from the purified α and β subunits, we performed an in vitro reassembly experiment for CPC and analyzed the reconstructed CPC using spectrophotometric and X-ray crystal structure analyses. The crystal structure of the reassembled CPC was nearly identical to that of the original CPC O77 . The findings of this study indicate that the octameric CPC O77 is a naturally occurring form in the thermophilic cyanobacterium Thermoleptolyngbya sp. O-77.
- Department of Chemistry and Biochemistry, Graduate School of Engineering, Kyushu University, 744 Moto-oka, Nishi-ku, Fukuoka 819-0395, Japan.
Organizational Affiliation: